Single-cell transcriptomics uncovers a non-autonomous Tbx1-dependent genetic program controlling cardiac neural crest cell development
- PMID: 36941249
- PMCID: PMC10027855
- DOI: 10.1038/s41467-023-37015-9
Single-cell transcriptomics uncovers a non-autonomous Tbx1-dependent genetic program controlling cardiac neural crest cell development
Abstract
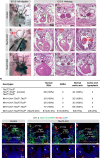
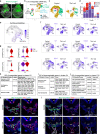
Disruption of cardiac neural crest cells (CNCCs) results in congenital heart disease, yet we do not understand the cell fate dynamics as these cells differentiate to vascular smooth muscle cells. Here we performed single-cell RNA-sequencing of NCCs from the pharyngeal apparatus with the heart in control mouse embryos and when Tbx1, the gene for 22q11.2 deletion syndrome, is inactivated. We uncover three dynamic transitions of pharyngeal NCCs expressing Tbx2 and Tbx3 through differentiated CNCCs expressing cardiac transcription factors with smooth muscle genes. These transitions are altered non-autonomously by loss of Tbx1. Further, inactivation of Tbx2 and Tbx3 in early CNCCs results in aortic arch branching defects due to failed smooth muscle differentiation. Loss of Tbx1 interrupts mesoderm to CNCC cell-cell communication with upregulation and premature activation of BMP signaling and reduced MAPK signaling, as well as alteration of other signaling, and failed dynamic transitions of CNCCs leading to disruption of aortic arch artery formation and cardiac outflow tract septation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

