Digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors within lipid bilayers
- PMID: 36941256
- PMCID: PMC10027858
- DOI: 10.1038/s41467-023-36996-x
Digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors within lipid bilayers
Abstract
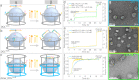
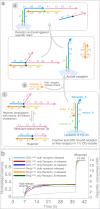
Interactions between membrane proteins are essential for cell survival but are often poorly understood. Even the biologically functional ratio of components within a multi-subunit membrane complex-the native stoichiometry-is difficult to establish. Here we demonstrate digital nanoreactors that can control interactions between lipid-bound molecular receptors along three key dimensions: stoichiometric, spatial, and temporal. Each nanoreactor is based on a DNA origami ring, which both templates the synthesis of a liposome and provides tethering sites for DNA-based receptors (modelling membrane proteins). Receptors are released into the liposomal membrane using strand displacement and a DNA logic gate measures receptor heterodimer formation. High-efficiency tethering of receptors enables the kinetics of receptors in 1:1 and 2:2 absolute stoichiometries to be observed by bulk fluorescence, which in principle is generalizable to any ratio. Similar single-molecule-in-bulk experiments using DNA-linked membrane proteins could determine native stoichiometry and the kinetics of membrane protein interactions for applications ranging from signalling research to drug discovery.
© 2023. The Author(s).
Conflict of interest statement
Certain aspects related to the internal immobilization and the release of membrane receptors employing DNA origami are covered in a pending US provisional patent application (63/415,546) owned by the California Institute of Technology with inventor V.M. The other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

