Small molecule metabolites: discovery of biomarkers and therapeutic targets
- PMID: 36941259
- PMCID: PMC10026263
- DOI: 10.1038/s41392-023-01399-3
Small molecule metabolites: discovery of biomarkers and therapeutic targets
Abstract
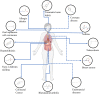
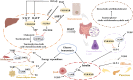
Metabolic abnormalities lead to the dysfunction of metabolic pathways and metabolite accumulation or deficiency which is well-recognized hallmarks of diseases. Metabolite signatures that have close proximity to subject's phenotypic informative dimension, are useful for predicting diagnosis and prognosis of diseases as well as monitoring treatments. The lack of early biomarkers could lead to poor diagnosis and serious outcomes. Therefore, noninvasive diagnosis and monitoring methods with high specificity and selectivity are desperately needed. Small molecule metabolites-based metabolomics has become a specialized tool for metabolic biomarker and pathway analysis, for revealing possible mechanisms of human various diseases and deciphering therapeutic potentials. It could help identify functional biomarkers related to phenotypic variation and delineate biochemical pathways changes as early indicators of pathological dysfunction and damage prior to disease development. Recently, scientists have established a large number of metabolic profiles to reveal the underlying mechanisms and metabolic networks for therapeutic target exploration in biomedicine. This review summarized the metabolic analysis on the potential value of small-molecule candidate metabolites as biomarkers with clinical events, which may lead to better diagnosis, prognosis, drug screening and treatment. We also discuss challenges that need to be addressed to fuel the next wave of breakthroughs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

