Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome
- PMID: 36941263
- PMCID: PMC10027818
- DOI: 10.1038/s41526-023-00247-6
Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome
Abstract
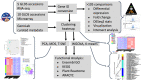
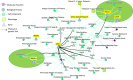
Spaceflight presents a multifaceted environment for plants, combining the effects on growth of many stressors and factors including altered gravity, the influence of experiment hardware, and increased radiation exposure. To help understand the plant response to this complex suite of factors this study compared transcriptomic analysis of 15 Arabidopsis thaliana spaceflight experiments deposited in the National Aeronautics and Space Administration's GeneLab data repository. These data were reanalyzed for genes showing significant differential expression in spaceflight versus ground controls using a single common computational pipeline for either the microarray or the RNA-seq datasets. Such a standardized approach to analysis should greatly increase the robustness of comparisons made between datasets. This analysis was coupled with extensive cross-referencing to a curated matrix of metadata associated with these experiments. Our study reveals that factors such as analysis type (i.e., microarray versus RNA-seq) or environmental and hardware conditions have important confounding effects on comparisons seeking to define plant reactions to spaceflight. The metadata matrix allows selection of studies with high similarity scores, i.e., that share multiple elements of experimental design, such as plant age or flight hardware. Comparisons between these studies then helps reduce the complexity in drawing conclusions arising from comparisons made between experiments with very different designs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Grants and funding
- 80NSSC19K0126/National Aeronautics and Space Administration (NASA)
- 80NSSC18K0132/National Aeronautics and Space Administration (NASA)
- 80NSSC21K0577/National Aeronautics and Space Administration (NASA)
- 80NSSC18K0132/National Aeronautics and Space Administration (NASA)
- 80NSSC19K0126/National Aeronautics and Space Administration (NASA)
- NNX15AG56G/National Aeronautics and Space Administration (NASA)
- NNX15AG56G/National Aeronautics and Space Administration (NASA)
- NNX15AG55G/National Aeronautics and Space Administration (NASA)
- 80NSSC19K1481/National Aeronautics and Space Administration (NASA)
- RTI2018-099309-B-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- 1340112 4000131202/20/NL/PG/pt/European Space Agency (ESA)
- 1127112/NSF | Directorate for Biological Sciences (BIO)
- 1127112/NSF | Directorate for Biological Sciences (BIO)
- 1340112/NSF | Directorate for Biological Sciences (BIO)
- 1340112/NSF | BIO | Division of Biological Infrastructure (DBI)
LinkOut - more resources
Full Text Sources

