Loss of H3K9 trimethylation alters chromosome compaction and transcription factor retention during mitosis
- PMID: 36941433
- PMCID: PMC10113154
- DOI: 10.1038/s41594-023-00943-7
Loss of H3K9 trimethylation alters chromosome compaction and transcription factor retention during mitosis
Abstract
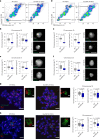
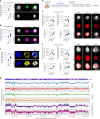
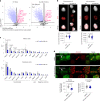
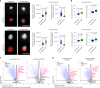
Recent studies have shown that repressive chromatin machinery, including DNA methyltransferases and polycomb repressor complexes, binds to chromosomes throughout mitosis and their depletion results in increased chromosome size. In the present study, we show that enzymes that catalyze H3K9 methylation, such as Suv39h1, Suv39h2, G9a and Glp, are also retained on mitotic chromosomes. Surprisingly, however, mutants lacking histone 3 lysine 9 trimethylation (H3K9me3) have unusually small and compact mitotic chromosomes associated with increased histone H3 phospho Ser10 (H3S10ph) and H3K27me3 levels. Chromosome size and centromere compaction in these mutants were rescued by providing exogenous first protein lysine methyltransferase Suv39h1 or inhibiting Ezh2 activity. Quantitative proteomic comparisons of native mitotic chromosomes isolated from wild-type versus Suv39h1/Suv39h2 double-null mouse embryonic stem cells revealed that H3K9me3 was essential for the efficient retention of bookmarking factors such as Esrrb. These results highlight an unexpected role for repressive heterochromatin domains in preserving transcription factor binding through mitosis and underscore the importance of H3K9me3 for sustaining chromosome architecture and epigenetic memory during cell division.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

