UniProt Tools: BLAST, Align, Peptide Search, and ID Mapping
- PMID: 36943033
- PMCID: PMC10034637
- DOI: 10.1002/cpz1.697
UniProt Tools: BLAST, Align, Peptide Search, and ID Mapping
Abstract
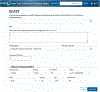
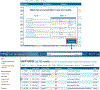
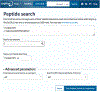
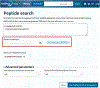
The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and annotation data (UniProt Consortium, 2023). The UniProt website receives about 800,000 unique visitors per month and is the primary means to access UniProt. Along with various datasets that you can search, UniProt provides four main tools. These are the "BLAST" tool for sequence similarity searching, the "Align" tool for multiple sequence alignment, the "Peptide Search" tool for retrieving proteins containing a short peptide sequence, and the "Retrieve/ID Mapping" tool for using a list of identifiers to retrieve UniProt Knowledgebase (UniProtKB) proteins and to convert database identifiers from UniProt to external databases or vice versa. This article provides four basic protocols and seven alternate protocols for using UniProt tools. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Basic local alignment search tool (BLAST) in UniProt Alternate Protocol 1: BLAST through UniProt text search results pages Alternate Protocol 2: BLAST through UniProt basket Basic Protocol 2: Multiple sequence alignment in UniProt Alternate Protocol 3: Align tool through UniProt results pages and entry pages Alternate Protocol 4: Align tool through UniProt basket Basic Protocol 3: Peptide search in UniProt Basic Protocol 4: Batch retrieval and ID mapping in UniProt Alternate Protocol 5: Retrieve/ID Mapping tool through UniProt text search results pages and BLAST and Align results pages Alternate Protocol 6: Retrieve/ID Mapping tool through UniProt basket Alternate Protocol 7: Retrieve/ID Mapping tool through UniProt search box.
Keywords: BLAST; UniProt; alignment; navigation; search; tutorial.
© 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures




























References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

