A Standardized Clinical Data Harmonization Pipeline for Scalable AI Application Deployment (FHIR-DHP): Validation and Usability Study
- PMID: 36943344
- PMCID: PMC10131740
- DOI: 10.2196/43847
A Standardized Clinical Data Harmonization Pipeline for Scalable AI Application Deployment (FHIR-DHP): Validation and Usability Study
Abstract
Background: Increasing digitalization in the medical domain gives rise to large amounts of health care data, which has the potential to expand clinical knowledge and transform patient care if leveraged through artificial intelligence (AI). Yet, big data and AI oftentimes cannot unlock their full potential at scale, owing to nonstandardized data formats, lack of technical and semantic data interoperability, and limited cooperation between stakeholders in the health care system. Despite the existence of standardized data formats for the medical domain, such as Fast Healthcare Interoperability Resources (FHIR), their prevalence and usability for AI remain limited.
Objective: In this paper, we developed a data harmonization pipeline (DHP) for clinical data sets relying on the common FHIR data standard.
Methods: We validated the performance and usability of our FHIR-DHP with data from the Medical Information Mart for Intensive Care IV database.
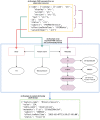
Results: We present the FHIR-DHP workflow in respect of the transformation of "raw" hospital records into a harmonized, AI-friendly data representation. The pipeline consists of the following 5 key preprocessing steps: querying of data from hospital database, FHIR mapping, syntactic validation, transfer of harmonized data into the patient-model database, and export of data in an AI-friendly format for further medical applications. A detailed example of FHIR-DHP execution was presented for clinical diagnoses records.
Conclusions: Our approach enables the scalable and needs-driven data modeling of large and heterogenous clinical data sets. The FHIR-DHP is a pivotal step toward increasing cooperation, interoperability, and quality of patient care in the clinical routine and for medical research.
Keywords: AI; AI application; FHIR; MIMIC IV; artificial intelligence; care; care unit; cooperation; data; data interoperability; data standardization pipeline; deployment; diagnosis; fast healthcare interoperability resources; medical information mart for intensive care; medical research; patient care; usability.
©Elena Williams, Manuel Kienast, Evelyn Medawar, Janis Reinelt, Alberto Merola, Sophie Anne Ines Klopfenstein, Anne Rike Flint, Patrick Heeren, Akira-Sebastian Poncette, Felix Balzer, Julian Beimes, Paul von Bünau, Jonas Chromik, Bert Arnrich, Nico Scherf, Sebastian Niehaus. Originally published in JMIR Medical Informatics (https://medinform.jmir.org), 21.03.2023.
Conflict of interest statement
Conflicts of Interest: FB reports grants from the German Federal Ministry of Education and Research, German Federal Ministry of Health, Berlin Institute of Health, personal fees from Elsevier Publishing, grants from Hans Böckler Foundation, other from Robert Koch Institute, grants from Einstein Foundation, grants from Berlin University Alliance, personal fees from Medtronic and personal fees from GE Healthcare.
Figures

References
-
- Desautels T, Calvert J, Hoffman J, Jay M, Kerem Y, Shieh L, Shimabukuro D, Chettipally U, Feldman MD, Barton C, Wales DJ, Das R. Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR Med Inform. 2016 Sep 30;4(3):e28. doi: 10.2196/medinform.5909. https://medinform.jmir.org/2016/3/e28/ v4i3e28 - DOI - PMC - PubMed
-
- Maier C, Kapsner LA, Mate S, Prokosch H, Kraus S. Patient Cohort Identification on Time Series Data Using the OMOP Common Data Model. Appl Clin Inform. 2021 Jan 27;12(1):57–64. doi: 10.1055/s-0040-1721481. http://www.thieme-connect.com/DOI/DOI?10.1055/s-0040-1721481 - DOI - PMC - PubMed
-
- Ni Y, Wright J, Perentesis J, Lingren T, Deleger L, Kaiser M, Kohane I, Solti I. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015 Apr 14;15(1):28. doi: 10.1186/s12911-015-0149-3. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-0... 10.1186/s12911-015-0149-3 - DOI - DOI - PMC - PubMed
-
- de Mello BH, Rigo SJ, da Costa CA, da Rosa Righi R, Donida B, Bez MR, Schunke LC. Semantic interoperability in health records standards: a systematic literature review. Health Technol (Berl) 2022;12(2):255–272. doi: 10.1007/s12553-022-00639-w. https://europepmc.org/abstract/MED/35103230 639 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

