Neutrophil Activation and Immune Thrombosis Profiles Persist in Convalescent COVID-19
- PMID: 36943669
- PMCID: PMC10029801
- DOI: 10.1007/s10875-023-01459-x
Neutrophil Activation and Immune Thrombosis Profiles Persist in Convalescent COVID-19
Erratum in
-
Correction to: Neutrophil Activation and Immune Thrombosis Profiles Persist in Convalescent COVID‑19.J Clin Immunol. 2023 Jul;43(5):894. doi: 10.1007/s10875-023-01477-9. J Clin Immunol. 2023. PMID: 36991251 Free PMC article. No abstract available.
Abstract
Purpose: Following a severe COVID-19 infection, a proportion of individuals develop prolonged symptoms. We investigated the immunological dysfunction that underlies the persistence of symptoms months after the resolution of acute COVID-19.
Methods: We analyzed cytokines, cell phenotypes, SARS-CoV-2 spike-specific and neutralizing antibodies, and whole blood gene expression profiles in convalescent severe COVID-19 patients 1, 3, and 6 months following hospital discharge.
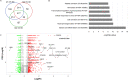
Results: We observed persistent abnormalities until month 6 marked by (i) high serum levels of monocyte/macrophage and endothelial activation markers, chemotaxis, and hematopoietic cytokines; (ii) a high frequency of central memory CD4+ and effector CD8+ T cells; (iii) a decrease in anti-SARS-CoV-2 spike and neutralizing antibodies; and (iv) an upregulation of genes related to platelet, neutrophil activation, erythrocytes, myeloid cell differentiation, and RUNX1 signaling. We identified a "core gene signature" associated with a history of thrombotic events, with upregulation of a set of genes involved in neutrophil activation, platelet, hematopoiesis, and blood coagulation.
Conclusion: The lack of restoration of gene expression to a normal profile after up to 6 months of follow-up, even in asymptomatic patients who experienced severe COVID-19, signals the need to carefully extend their clinical follow-up and propose preventive measures.
Trial registration: ClinicalTrials.gov NCT04262921.
Keywords: COVID-19 disease; post-acute COVID-19 syndrome; thrombosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

