The Long Road to a Synthetic Self-Replicating Central Dogma
- PMID: 36944355
- PMCID: PMC10077596
- DOI: 10.1021/acs.biochem.3c00023
The Long Road to a Synthetic Self-Replicating Central Dogma
Abstract
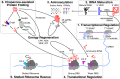
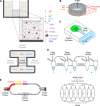
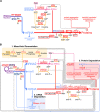
The construction of a biochemical system capable of self-replication is a key objective in bottom-up synthetic biology. Throughout the past two decades, a rapid progression in the design of in vitro cell-free systems has provided valuable insight into the requirements for the development of a minimal system capable of self-replication. The main limitations of current systems can be attributed to their macromolecular composition and how the individual macromolecules use the small molecules necessary to drive RNA and protein synthesis. In this Perspective, we discuss the recent steps that have been taken to generate a minimal cell-free system capable of regenerating its own macromolecular components and maintaining the homeostatic balance between macromolecular biogenesis and consumption of primary building blocks. By following the flow of biological information through the central dogma, we compare the current versions of these systems to date and propose potential alterations aimed at designing a model system for self-replicative synthetic cells.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

