Pan-genome inversion index reveals evolutionary insights into the subpopulation structure of Asian rice
- PMID: 36944612
- PMCID: PMC10030860
- DOI: 10.1038/s41467-023-37004-y
Pan-genome inversion index reveals evolutionary insights into the subpopulation structure of Asian rice
Abstract
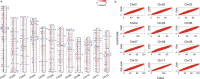
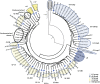
Understanding and exploiting genetic diversity is a key factor for the productive and stable production of rice. Here, we utilize 73 high-quality genomes that encompass the subpopulation structure of Asian rice (Oryza sativa), plus the genomes of two wild relatives (O. rufipogon and O. punctata), to build a pan-genome inversion index of 1769 non-redundant inversions that span an average of ~29% of the O. sativa cv. Nipponbare reference genome sequence. Using this index, we estimate an inversion rate of ~700 inversions per million years in Asian rice, which is 16 to 50 times higher than previously estimated for plants. Detailed analyses of these inversions show evidence of their effects on gene expression, recombination rate, and linkage disequilibrium. Our study uncovers the prevalence and scale of large inversions (≥100 bp) across the pan-genome of Asian rice and hints at their largely unexplored role in functional biology and crop performance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

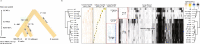



References
-
- Hossain M, Fischer K. Rice research for food security and sustainable agricultural development in Asia: achievements and future challenges. GeoJournal. 1995;35:286–298. doi: 10.1007/BF00989136. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

