A symbiotic physical niche in Drosophila melanogaster regulates stable association of a multi-species gut microbiota
- PMID: 36944617
- PMCID: PMC10030875
- DOI: 10.1038/s41467-023-36942-x
A symbiotic physical niche in Drosophila melanogaster regulates stable association of a multi-species gut microbiota
Abstract
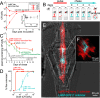
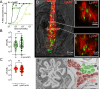
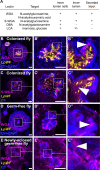
The gut is continuously invaded by diverse bacteria from the diet and the environment, yet microbiome composition is relatively stable over time for host species ranging from mammals to insects, suggesting host-specific factors may selectively maintain key species of bacteria. To investigate host specificity, we used gnotobiotic Drosophila, microbial pulse-chase protocols, and microscopy to investigate the stability of different strains of bacteria in the fly gut. We show that a host-constructed physical niche in the foregut selectively binds bacteria with strain-level specificity, stabilizing their colonization. Primary colonizers saturate the niche and exclude secondary colonizers of the same strain, but initial colonization by Lactobacillus species physically remodels the niche through production of a glycan-rich secretion to favor secondary colonization by unrelated commensals in the Acetobacter genus. Our results provide a mechanistic framework for understanding the establishment and stability of a multi-species intestinal microbiome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

