Chromatin remodeler Activity-Dependent Neuroprotective Protein (ADNP) contributes to syndromic autism
- PMID: 36945042
- PMCID: PMC10031977
- DOI: 10.1186/s13148-023-01450-8
Chromatin remodeler Activity-Dependent Neuroprotective Protein (ADNP) contributes to syndromic autism
Abstract
Background: Individuals affected with autism often suffer additional co-morbidities such as intellectual disability. The genes contributing to autism cluster on a relatively limited number of cellular pathways, including chromatin remodeling. However, limited information is available on how mutations in single genes can result in such pleiotropic clinical features in affected individuals. In this review, we summarize available information on one of the most frequently mutated genes in syndromic autism the Activity-Dependent Neuroprotective Protein (ADNP).
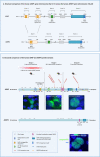
Results: Heterozygous and predicted loss-of-function ADNP mutations in individuals inevitably result in the clinical presentation with the Helsmoortel-Van der Aa syndrome, a frequent form of syndromic autism. ADNP, a zinc finger DNA-binding protein has a role in chromatin remodeling: The protein is associated with the pericentromeric protein HP1, the SWI/SNF core complex protein BRG1, and other members of this chromatin remodeling complex and, in murine stem cells, with the chromodomain helicase CHD4 in a ChAHP complex. ADNP has recently been shown to possess R-loop processing activity. In addition, many additional functions, for instance, in association with cytoskeletal proteins have been linked to ADNP.
Conclusions: We here present an integrated evaluation of all current aspects of gene function and evaluate how abnormalities in chromatin remodeling might relate to the pleiotropic clinical presentation in individual"s" with Helsmoortel-Van der Aa syndrome.
Keywords: ADNP; Activity-Dependent Neuroprotective Protein; Autism, intellectual disability, neurodevelopmental disorder, cancer; Chromatin remodeler; Helsmoortel–Van der Aa syndrome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest. Professor Illana Gozes is a Co-Founder, Chief Scientific Officer and a Director at ATED Therapeutics, clinically developing davunetide.
Figures


References
-
- American Psychiatric Association. American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders. Fifth. Arlington, VA; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

