This is a preprint.
Roles of microbiota in autoimmunity in Arabidopsis
- PMID: 36945461
- PMCID: PMC10028843
- DOI: 10.1101/2023.03.06.531303
Roles of microbiota in autoimmunity in Arabidopsis
Update in
-
Roles of microbiota in autoimmunity in Arabidopsis leaves.Nat Plants. 2024 Sep;10(9):1363-1376. doi: 10.1038/s41477-024-01779-9. Epub 2024 Sep 6. Nat Plants. 2024. PMID: 39242981 Free PMC article.
Abstract
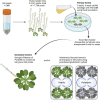
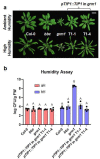
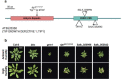
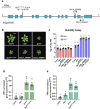
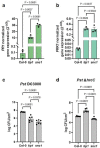
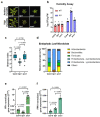
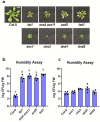
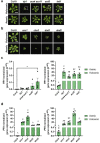
Over the past three decades, researchers have isolated plant mutants that display constitutively activated defense responses in the absence of pathogen infection. These mutants are called autoimmune mutants and are typically dwarf and/or bearing chlorotic/necrotic lesions. From a genetic screen for Arabidopsis genes involved in maintaining a normal leaf microbiota, we identified TIP GROWTH DEFECTIVE 1 (TIP1), which encodes a S-acyltransferase, as a key player in guarding leaves against abnormal microbiota level and composition under high humidity conditions. The tip1 mutant has several characteristic phenotypes of classical autoimmune mutants, including a dwarf stature, displaying lesions, and having a high basal level of defense gene expression. Gnotobiotic experiments revealed that the autoimmune phenotypes of the tip1 mutant are largely dependent on the presence of microbiota as axenic tip1 plants have markedly reduced autoimmune phenotypes. We found that the microbiota dependency of autoimmune phenotypes is shared by several "lesion mimic"-type autoimmune mutants in Arabidopsis. Interestingly, autoimmune phenotypes caused by mutations in NLR genes do not require the presence of microbiota and can even be partially alleviated by microbiota. Our results therefore suggest the existence of two classes of autoimmunity (microbiota-dependent vs. microbiota-independent) in plants. The observed interplay between autoimmunity and microbiota in the lesion mimic class of autoimmunity is reminiscent of the interactions between autoimmunity and dysbiosis in the animal kingdom.
Conflict of interest statement
Ethics declarations Competing interests The authors declare no competing interests.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
