This is a preprint.
T-CLEARE: A Pilot Community-Driven Tissue-Clearing Protocol Repository
- PMID: 36945489
- PMCID: PMC10028991
- DOI: 10.1101/2023.03.09.531970
T-CLEARE: A Pilot Community-Driven Tissue-Clearing Protocol Repository
Update in
-
T-CLEARE: a pilot community-driven tissue clearing protocol repository.Front Bioeng Biotechnol. 2024 Sep 16;12:1304622. doi: 10.3389/fbioe.2024.1304622. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39351064 Free PMC article.
Abstract
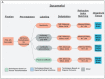
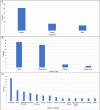
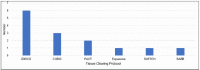
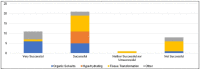
Selecting and implementing a tissue-clearing protocol is challenging. Established more than 100 years ago, tissue clearing is still a rapidly evolving field of research. There are currently many published protocols to choose from, and each performs better or worse across a range of key evaluation factors (e.g., speed, cost, tissue stability, fluorescence quenching). Additionally, tissue-clearing protocols are often optimized for specific experimental contexts, and applying an existing protocol to a new problem can require a lengthy period of adaptation by trial and error. Although the primary literature and review articles provide a useful starting point for optimization, there is growing recognition that many articles do not provide sufficient detail to replicate or reproduce experimental results. To help address this issue, we have developed a novel, freely available repository of tissue-clearing protocols named T-CLEARE (Tissue CLEAring protocol REpository; https://doryworkspace.org/doryviz). T-CLEARE incorporates community responses to an open survey designed to capture details not commonly found in the scientific literature, including modifications to published protocols required for specific use cases and instances when tissue-clearing protocols did not perform well (negative results). The goal of T-CLEARE is to provide a forum for the community to share evaluations and modifications of tissue-clearing protocols for various tissue types and potentially identify best-in-class methods for a given application.
Figures



References
-
- Chhetri RK, Amat F, Wan Y, Hockendorf B, Lemon WC, Keller PJ. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods. 2015;12(12):1171–8. - PubMed
-
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10(5):413–20. - PubMed
-
- Vladimirov N, Mu Y, Kawashima T, Bennett DV, Yang CT, Looger LL, et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat Methods. 2014;11(9):883–4. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
