This is a preprint.
Dissecting embryonic and extra-embryonic lineage crosstalk with stem cell co-culture
- PMID: 36945498
- PMCID: PMC10028955
- DOI: 10.1101/2023.03.07.531525
Dissecting embryonic and extra-embryonic lineage crosstalk with stem cell co-culture
Update in
-
Dissecting embryonic and extraembryonic lineage crosstalk with stem cell co-culture.Cell. 2023 Dec 21;186(26):5859-5875.e24. doi: 10.1016/j.cell.2023.11.008. Epub 2023 Dec 4. Cell. 2023. PMID: 38052213 Free PMC article.
Abstract
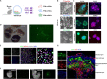
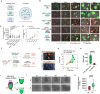
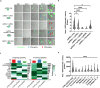
Faithful embryogenesis requires precise coordination between embryonic and extraembryonic tissues. Although stem cells from embryonic and extraembryonic origins have been generated for several mammalian species(Bogliotti et al., 2018; Choi et al., 2019; Cui et al., 2019; Evans and Kaufman, 1981; Kunath et al., 2005; Li et al., 2008; Martin, 1981; Okae et al., 2018; Tanaka et al., 1998; Thomson et al., 1998; Vandevoort et al., 2007; Vilarino et al., 2020; Yu et al., 2021b; Zhong et al., 2018), they are grown in different culture conditions with diverse media composition, which makes it difficult to study cross-lineage communication. Here, by using the same culture condition that activates FGF, TGF-β and WNT signaling pathways, we derived stable embryonic stem cells (ESCs), extraembryonic endoderm stem cells (XENs) and trophoblast stem cells (TSCs) from all three founding tissues of mouse and cynomolgus monkey blastocysts. This allowed us to establish embryonic and extraembryonic stem cell co-cultures to dissect lineage crosstalk during early mammalian development. Co-cultures of ESCs and XENs uncovered a conserved and previously unrecognized growth inhibition of pluripotent cells by extraembryonic endoderm cells, which is in part mediated through extracellular matrix signaling. Our study unveils a more universal state of stem cell self-renewal stabilized by activation, as opposed to inhibition, of developmental signaling pathways. The embryonic and extraembryonic stem cell co-culture strategy developed here will open new avenues for creating more faithful embryo models and developing more developmentally relevant differentiation protocols.
Conflict of interest statement
DECLARATION OF INTERESTS Y.W., L.Y., T.T. and J.W. are inventors on a patent application (applied through the Board of Regents of The University of Texas System, application number 63/488,401) entitled “Methods For the Derivation Culture Of Embryonic and Extra-Embryonic Stem Cells” arising from this work. The other authors declare no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
