This is a preprint.
Circulating tumor DNA association with residual cancer burden after neoadjuvant chemotherapy in triple-negative breast cancer in TBCRC 030
- PMID: 36945501
- PMCID: PMC10029037
- DOI: 10.1101/2023.03.06.23286772
Circulating tumor DNA association with residual cancer burden after neoadjuvant chemotherapy in triple-negative breast cancer in TBCRC 030
Update in
-
Circulating tumor DNA association with residual cancer burden after neoadjuvant chemotherapy in triple-negative breast cancer in TBCRC 030.Ann Oncol. 2023 Oct;34(10):899-906. doi: 10.1016/j.annonc.2023.08.004. Epub 2023 Aug 18. Ann Oncol. 2023. PMID: 37597579 Free PMC article.
Abstract
Purpose: To examine circulating tumor DNA (ctDNA) and its association with residual cancer burden (RCB) using an ultrasensitive assay in patients with triple-negative breast cancer (TNBC) receiving neoadjuvant chemotherapy (NAT).
Patients and methods: We identified responders (RCB-0/1) and matched non-responders (RCB-2/3) from the phase II TBCRC 030 prospective study of neoadjuvant paclitaxel vs. cisplatin in TNBC. We collected plasma samples at baseline, three weeks, and twelve weeks (end of therapy). We created personalized ctDNA assays utilizing MAESTRO mutation enrichment sequencing. We explored associations between ctDNA and RCB status and disease recurrence.
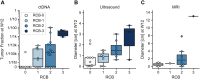
Results: Of 139 patients, 68 had complete samples and no additional NAT. Twenty-two were responders and 19 of those had sufficient tissue for whole-genome sequencing. We identified an additional 19 non-responders for a matched case-control analysis of 38 patients using a MAESTRO ctDNA assay tracking 319-1000 variants (median 1000) to 114 plasma samples from 3 timepoints. Overall, ctDNA positivity was 100% at baseline, 79% at week 3, and 55% at week 12. Median tumor fraction (TFx) was 3.7 × 10 -4 (range: 7.9 × 10 -7 to 4.9 × 10 -1 ). TFx decreased 285-fold from baseline to week 3 in responders and 24-fold in non-responders. Week 12 ctDNA clearance correlated with RCB: clearance was observed in 10/11 patients with RCB-0, 3/8 with RCB-1, 4/15 with RCB-2, and 0/4 with RCB-3. Among 6 patients with known recurrence five had persistent ctDNA at week 12.
Conclusion: NAT for TNBC reduced ctDNA TFx by 285-fold in responders and 24-fold in non-responders. In 58% (22/38) of patients, ctDNA TFx dropped below the detection level of a commercially available test, emphasizing the need for sensitive tests. Additional studies will determine if ctDNA-guided approaches can improve outcomes.
Conflict of interest statement
Conflicts of Interest:
All other authors report no disclosures.
Figures


References
-
- Foulkes WD, Smith IE, Reis-Filho JS: Triple-negative breast cancer. N Engl J Med 363:1938–48, 2010 - PubMed
-
- Liedtke C, Mazouni C, Hess KR, et al. : Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26:1275–81, 2008 - PubMed
-
- Dent R, Trudeau M, Pritchard KI, et al. : Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13:4429–34, 2007 - PubMed
-
- Haffty BG, Yang Q, Reiss M, et al. : Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 24:5652–7, 2006 - PubMed
-
- von Minckwitz G, Untch M, Blohmer JU, et al. : Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 30:1796–804, 2012 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
