Folding@home: Achievements from over 20 years of citizen science herald the exascale era
- PMID: 36945779
- PMCID: PMC10398258
- DOI: 10.1016/j.bpj.2023.03.028
Folding@home: Achievements from over 20 years of citizen science herald the exascale era
Abstract
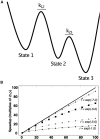
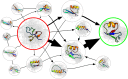
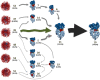
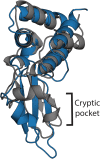
Simulations of biomolecules have enormous potential to inform our understanding of biology but require extremely demanding calculations. For over 20 years, the Folding@home distributed computing project has pioneered a massively parallel approach to biomolecular simulation, harnessing the resources of citizen scientists across the globe. Here, we summarize the scientific and technical advances this perspective has enabled. As the project's name implies, the early years of Folding@home focused on driving advances in our understanding of protein folding by developing statistical methods for capturing long-timescale processes and facilitating insight into complex dynamical processes. Success laid a foundation for broadening the scope of Folding@home to address other functionally relevant conformational changes, such as receptor signaling, enzyme dynamics, and ligand binding. Continued algorithmic advances, hardware developments such as graphics processing unit (GPU)-based computing, and the growing scale of Folding@home have enabled the project to focus on new areas where massively parallel sampling can be impactful. While previous work sought to expand toward larger proteins with slower conformational changes, new work focuses on large-scale comparative studies of different protein sequences and chemical compounds to better understand biology and inform the development of small-molecule drugs. Progress on these fronts enabled the community to pivot quickly in response to the COVID-19 pandemic, expanding to become the world's first exascale computer and deploying this massive resource to provide insight into the inner workings of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus and aid the development of new antivirals. This success provides a glimpse of what is to come as exascale supercomputers come online and as Folding@home continues its work.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests G.R.B. is a cofounder and board member of Decrypt Biomedicine. V.S.P. is the Managing Partner of a16z BioHealth and is deeply involved in a rapidly evolving set of companies.
Figures
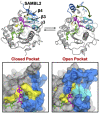
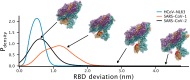
Update of
-
Folding@home: achievements from over twenty years of citizen science herald the exascale era.ArXiv [Preprint]. 2023 Mar 15:arXiv:2303.08993v1. ArXiv. 2023. Update in: Biophys J. 2023 Jul 25;122(14):2852-2863. doi: 10.1016/j.bpj.2023.03.028. PMID: 36994157 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

