Sexually antagonistic selection maintains genetic variance when sexual dimorphism evolves
- PMID: 36946115
- PMCID: PMC10031426
- DOI: 10.1098/rspb.2022.2484
Sexually antagonistic selection maintains genetic variance when sexual dimorphism evolves
Abstract
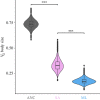
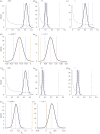
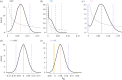
Genetic variance (VG) in fitness related traits is often unexpectedly high, evoking the question how VG can be maintained in the face of selection. Sexually antagonistic (SA) selection favouring alternative alleles in the sexes is common and predicted to maintain VG, while directional selection should erode it. Both SA and sex-limited directional selection can lead to sex-specific adaptations but how each affect VG when sexual dimorphism evolves remain experimentally untested. Using replicated artificial selection on the seed beetle Callosobruchus maculatus body size we recently demonstrated an increase in size dimorphism under SA and male-limited (ML) selection by 50% and 32%, respectively. Here we test their consequences on genetic variation. We show that SA selection maintained significantly more ancestral, autosomal additive genetic variance than ML selection, while both eroded sex-linked additive variation equally. Ancestral female-specific dominance variance was completely lost under ML, while SA selection consistently sustained it. Further, both forms of selection preserved a high genetic correlation between the sexes (rm,f). These results demonstrate the potential for sexual antagonism to maintain more genetic variance while fuelling sex-specific adaptation in a short evolutionary time scale, and are in line with predicted importance of sex-specific dominance reducing sexual conflict over alternative alleles.
Keywords: animal model; artificial selection; balancing selection; body size; dominance variation; sexual conflict.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
