Utility of targeted gene sequencing to differentiate myeloid malignancies from other cytopenic conditions
- PMID: 36947201
- PMCID: PMC10368770
- DOI: 10.1182/bloodadvances.2022008578
Utility of targeted gene sequencing to differentiate myeloid malignancies from other cytopenic conditions
Abstract
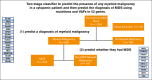
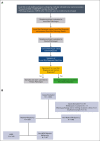
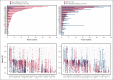
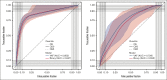
The National Heart, Lung, and Blood Institute-funded National MDS Natural History Study (NCT02775383) is a prospective cohort study enrolling patients with cytopenia with suspected myelodysplastic syndromes (MDS) to evaluate factors associated with disease. Here, we sequenced 53 genes in bone marrow samples harvested from 1298 patients diagnosed with myeloid malignancy, including MDS and non-MDS myeloid malignancy or alternative marrow conditions with cytopenia based on concordance between independent histopathologic reviews (local, centralized, and tertiary to adjudicate disagreements when needed). We developed a novel 2-stage diagnostic classifier based on mutational profiles in 18 of 53 sequenced genes that were sufficient to best predict a diagnosis of myeloid malignancy and among those with a predicted myeloid malignancy, predict whether they had MDS. The classifier achieved a positive predictive value (PPV) of 0.84 and negative predictive value (NPV) of 0.8 with an area under the receiver operating characteristic curve (AUROC) of 0.85 when classifying patients as having myeloid vs no myeloid malignancy based on variant allele frequencies (VAFs) in 17 genes and a PPV of 0.71 and NPV of 0.64 with an AUROC of 0.73 when classifying patients as having MDS vs non-MDS malignancy based on VAFs in 10 genes. We next assessed how this approach could complement histopathology to improve diagnostic accuracy. For 99 of 139 (71%) patients (PPV of 0.83 and NPV of 0.65) with local and centralized histopathologic disagreement in myeloid vs no myeloid malignancy, the classifier-predicted diagnosis agreed with the tertiary pathology review (considered the internal gold standard).
Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution.
Conflict of interest statement
Conflict-of-interest disclosure: A.E.D. reports consultancy and membership on a board or advisory committee for Taiho, Novartis, and Bristol Myers Squibb and membership on a board or advisory committee for Takeda. R.C.L. reports consultancy for Takeda, bluebird bio, and Thermo Fisher Scientific. R.B. reports employment and stock in private company of Aptose Biosciences; has a chair of data safety monitoring committee in Gilead and Epizyme; has membership on a board or advisory committee in Silence Therapeutics; and receives research funding from Takeda. T.A.B. has stock in private company and membership on a board or advisory committee in Bristol Myers Squibb; has stock in private company in AstraZeneca, Epizyme, and Heron Therapeutics; and has membership on a board or advisory committee in MorphoSys, Karyopharm, and Cardinal Health. J.M. has membership on a board or advisory committee in bone marrow failure. E.P. receives research funding from Bristol Myers Squibb, Kura, and Incyte and receives honoraria from Taiho and Blueprint. R.K. receives honoraria from Novartis, Geron, and Acceleron, and receives honoraria from and is on the speakers bureau of Agios, AbbVie, Jazz, and Bristol Myers Squibb. M.A.S. reports membership on a board or advisory committee for Novartis, Takeda/Millenium, and Bristol Myers Squibb. The remaining authors declare no competing financial interests.
Figures
References
-
- Galli A, Todisco G, Catamo E, et al. Relationship between clone metrics and clinical outcome in clonal cytopenia. Blood. 2021;138(11):965–976. - PubMed
-
- Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

