A real data-driven simulation strategy to select an imputation method for mixed-type trait data
- PMID: 36947561
- PMCID: PMC10069776
- DOI: 10.1371/journal.pcbi.1010154
A real data-driven simulation strategy to select an imputation method for mixed-type trait data
Abstract
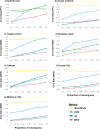
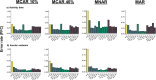
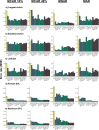
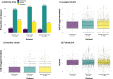
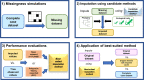
Missing observations in trait datasets pose an obstacle for analyses in myriad biological disciplines. Considering the mixed results of imputation, the wide variety of available methods, and the varied structure of real trait datasets, a framework for selecting a suitable imputation method is advantageous. We invoked a real data-driven simulation strategy to select an imputation method for a given mixed-type (categorical, count, continuous) target dataset. Candidate methods included mean/mode imputation, k-nearest neighbour, random forests, and multivariate imputation by chained equations (MICE). Using a trait dataset of squamates (lizards and amphisbaenians; order: Squamata) as a target dataset, a complete-case dataset consisting of species with nearly complete information was formed for the imputation method selection. Missing data were induced by removing values from this dataset under different missingness mechanisms: missing completely at random (MCAR), missing at random (MAR), and missing not at random (MNAR). For each method, combinations with and without phylogenetic information from single gene (nuclear and mitochondrial) or multigene trees were used to impute the missing values for five numerical and two categorical traits. The performances of the methods were evaluated under each missing mechanism by determining the mean squared error and proportion falsely classified rates for numerical and categorical traits, respectively. A random forest method supplemented with a nuclear-derived phylogeny resulted in the lowest error rates for the majority of traits, and this method was used to impute missing values in the original dataset. Data with imputed values better reflected the characteristics and distributions of the original data compared to complete-case data. However, caution should be taken when imputing trait data as phylogeny did not always improve performance for every trait and in every scenario. Ultimately, these results support the use of a real data-driven simulation strategy for selecting a suitable imputation method for a given mixed-type trait dataset.
Copyright: © 2023 May et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Valcu M, Dale J, Griesser M, Nakagawa S, Kempenaers B. Global gradients of avian longevity support the classic evolutionary theory of ageing. Ecography. 2014. Oct 1;37(10):930–8.
-
- Howard SD, Bickford DP. Amphibians over the edge: silent extinction risk of Data Deficient species. Divers Distrib. 2014. Jul 1;20(7):837–46.
-
- Pacifici M, Visconti P, Butchart SHM, Watson JEM, Cassola FM, Rondinini C. Species’ traits influenced their response to recent climate change. Nat Clim Change. 2017. Mar 1;7(3):205–8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

