Epigenetic dosage identifies two major and functionally distinct β cell subtypes
- PMID: 36948185
- PMCID: PMC10160009
- DOI: 10.1016/j.cmet.2023.03.008
Epigenetic dosage identifies two major and functionally distinct β cell subtypes
Abstract
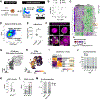
The mechanisms that specify and stabilize cell subtypes remain poorly understood. Here, we identify two major subtypes of pancreatic β cells based on histone mark heterogeneity (βHI and βLO). βHI cells exhibit ∼4-fold higher levels of H3K27me3, distinct chromatin organization and compaction, and a specific transcriptional pattern. βHI and βLO cells also differ in size, morphology, cytosolic and nuclear ultrastructure, epigenomes, cell surface marker expression, and function, and can be FACS separated into CD24+ and CD24- fractions. Functionally, βHI cells have increased mitochondrial mass, activity, and insulin secretion in vivo and ex vivo. Partial loss of function indicates that H3K27me3 dosage regulates βHI/βLO ratio in vivo, suggesting that control of β cell subtype identity and ratio is at least partially uncoupled. Both subtypes are conserved in humans, with βHI cells enriched in humans with type 2 diabetes. Thus, epigenetic dosage is a novel regulator of cell subtype specification and identifies two functionally distinct β cell subtypes.
Keywords: H3K27me3; beta cells; bivalent genes; cell heterogeneity; chromatin organization; epigenetics; insulin; pancreatic islet; single cell.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

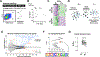




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

