A mechanism by which gut microbiota elevates permeability and inflammation in obese/diabetic mice and human gut
- PMID: 36948576
- PMCID: PMC10512000
- DOI: 10.1136/gutjnl-2022-327365
A mechanism by which gut microbiota elevates permeability and inflammation in obese/diabetic mice and human gut
Abstract
Objective: Ample evidence exists for the role of abnormal gut microbiota composition and increased gut permeability ('leaky gut') in chronic inflammation that commonly co-occurs in the gut in both obesity and diabetes, yet the detailed mechanisms involved in this process have remained elusive.
Design: In this study, we substantiate the causal role of the gut microbiota by use of faecal conditioned media along with faecal microbiota transplantation. Using untargeted and comprehensive approaches, we discovered the mechanism by which the obese microbiota instigates gut permeability, inflammation and abnormalities in glucose metabolism.
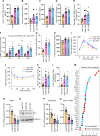
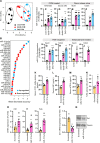
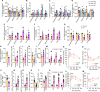
Results: We demonstrated that the reduced capacity of the microbiota from both obese mice and humans to metabolise ethanolamine results in ethanolamine accumulation in the gut, accounting for induction of intestinal permeability. Elevated ethanolamine increased the expression of microRNA-miR-101a-3p by enhancing ARID3a binding on the miR promoter. Increased miR-101a-3p decreased the stability of zona occludens-1 (Zo1) mRNA, which in turn, weakened intestinal barriers and induced gut permeability, inflammation and abnormalities in glucose metabolism. Importantly, restoring ethanolamine-metabolising activity in gut microbiota using a novel probiotic therapy reduced elevated gut permeability, inflammation and abnormalities in glucose metabolism by correcting the ARID3a/miR-101a/Zo1 axis.
Conclusion: Overall, we discovered that the reduced capacity of obese microbiota to metabolise ethanolamine instigates gut permeability, inflammation and glucose metabolic dysfunctions, and restoring ethanolamine-metabolising capacity by a novel probiotic therapy reverses these abnormalities.
Trial registration number: NCT02869659 and NCT03269032.
Keywords: diabetes mellitus; gut inflammation; inflammation; intestinal microbiology; obesity.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: The intellectual property for HL-200 probiotics is under-review with institutional patent and technology office. HY is a chief scientific officer of the Postbiotics, which has no influence and contribution with the work done in current manuscript.
Figures





Comment in
-
Obesity promotes a leaky gut, inflammation and pre-diabetes by lowering gut microbiota that metabolise ethanolamine.Gut. 2023 Oct;72(10):1809-1811. doi: 10.1136/gutjnl-2023-329815. Epub 2023 Apr 27. Gut. 2023. PMID: 37105722 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
