Checkpoint kinase 1/2 inhibition potentiates anti-tumoral immune response and sensitizes gliomas to immune checkpoint blockade
- PMID: 36949040
- PMCID: PMC10033639
- DOI: 10.1038/s41467-023-36878-2
Checkpoint kinase 1/2 inhibition potentiates anti-tumoral immune response and sensitizes gliomas to immune checkpoint blockade
Abstract
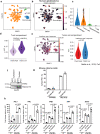
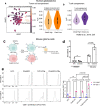
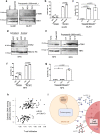
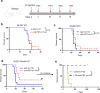
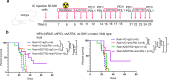
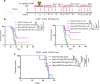
Whereas the contribution of tumor microenvironment to the profound immune suppression of glioblastoma (GBM) is clear, tumor-cell intrinsic mechanisms that regulate resistance to CD8 T cell mediated killing are less understood. Kinases are potentially druggable targets that drive tumor progression and might influence immune response. Here, we perform an in vivo CRISPR screen to identify glioma intrinsic kinases that contribute to evasion of tumor cells from CD8 T cell recognition. The screen reveals checkpoint kinase 2 (Chek2) to be the most important kinase contributing to escape from CD8 T-cell recognition. Genetic depletion or pharmacological inhibition of Chek2 with blood-brain-barrier permeable drugs that are currently being evaluated in clinical trials, in combination with PD-1 or PD-L1 blockade, lead to survival benefit in multiple preclinical glioma models. Mechanistically, loss of Chek2 enhances antigen presentation, STING pathway activation and PD-L1 expression in mouse gliomas. Analysis of human GBMs demonstrates that Chek2 expression is inversely associated with antigen presentation and T-cell activation. Collectively, these results support Chek2 as a promising target for enhancement of response to immune checkpoint blockade therapy in GBM.
© 2023. The Author(s).
Conflict of interest statement
C.D., A.M.S., C.L-C, and L.C. are co-authors for the following patent filed by Northwestern University: Method of using checkpoint kinase 1/2 inhibitor therapy to modulate anti-tumoral response against cancer and sensitize gliomas to immunotherapy (US Patent App. 16/951,638). A.M.S. has received in-kind and or funding support for research from Agenus, BMS, and Carthera. R.R. is a member of the SAB of AimedBio, consultant for Arquimea Research and a founder of Genotwin. The remaining authors declare no other competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

