YBX1 integration of oncogenic PI3K/mTOR signalling regulates the fitness of malignant epithelial cells
- PMID: 36949044
- PMCID: PMC10033729
- DOI: 10.1038/s41467-023-37161-0
YBX1 integration of oncogenic PI3K/mTOR signalling regulates the fitness of malignant epithelial cells
Abstract
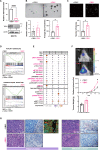
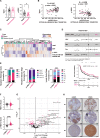
In heterogeneous head and neck cancer (HNC), subtype-specific treatment regimens are currently missing. An integrated analysis of patient HNC subtypes using single-cell sequencing and proteome profiles reveals an epithelial-mesenchymal transition (EMT) signature within the epithelial cancer-cell population. The EMT signature coincides with PI3K/mTOR inactivation in the mesenchymal subtype. Conversely, the signature is suppressed in epithelial cells of the basal subtype which exhibits hyperactive PI3K/mTOR signalling. We further identify YBX1 phosphorylation, downstream of the PI3K/mTOR pathway, restraining basal-like cancer cell proliferation. In contrast, YBX1 acts as a safeguard against the proliferation-to-invasion switch in mesenchymal-like epithelial cancer cells, and its loss accentuates partial-EMT and in vivo invasion. Interestingly, phospho-YBX1 that is mutually exclusive to partial-EMT, emerges as a prognostic marker for overall patient outcomes. These findings create a unique opportunity to sensitise mesenchymal cancer cells to PI3K/mTOR inhibitors by shifting them towards a basal-like subtype as a promising therapeutic approach against HNC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

