To metabolomics and beyond: a technological portfolio to investigate cancer metabolism
- PMID: 36949046
- PMCID: PMC10033890
- DOI: 10.1038/s41392-023-01380-0
To metabolomics and beyond: a technological portfolio to investigate cancer metabolism
Abstract
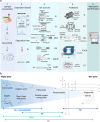
Tumour cells have exquisite flexibility in reprogramming their metabolism in order to support tumour initiation, progression, metastasis and resistance to therapies. These reprogrammed activities include a complete rewiring of the bioenergetic, biosynthetic and redox status to sustain the increased energetic demand of the cells. Over the last decades, the cancer metabolism field has seen an explosion of new biochemical technologies giving more tools than ever before to navigate this complexity. Within a cell or a tissue, the metabolites constitute the direct signature of the molecular phenotype and thus their profiling has concrete clinical applications in oncology. Metabolomics and fluxomics, are key technological approaches that mainly revolutionized the field enabling researchers to have both a qualitative and mechanistic model of the biochemical activities in cancer. Furthermore, the upgrade from bulk to single-cell analysis technologies provided unprecedented opportunity to investigate cancer biology at cellular resolution allowing an in depth quantitative analysis of complex and heterogenous diseases. More recently, the advent of functional genomic screening allowed the identification of molecular pathways, cellular processes, biomarkers and novel therapeutic targets that in concert with other technologies allow patient stratification and identification of new treatment regimens. This review is intended to be a guide for researchers to cancer metabolism, highlighting current and emerging technologies, emphasizing advantages, disadvantages and applications with the potential of leading the development of innovative anti-cancer therapies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Fendt SM, Frezza C, Erez A. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10:1797–1807. doi: 10.1158/2159-8290.CD-20-0844. - DOI - PMC - PubMed
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

