A DNA tumor virus globally reprograms host 3D genome architecture to achieve immortal growth
- PMID: 36949074
- PMCID: PMC10033825
- DOI: 10.1038/s41467-023-37347-6
A DNA tumor virus globally reprograms host 3D genome architecture to achieve immortal growth
Abstract
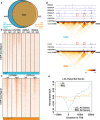
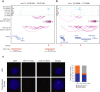
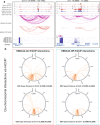
Epstein-Barr virus (EBV) immortalization of resting B lymphocytes (RBLs) to lymphoblastoid cell lines (LCLs) models human DNA tumor virus oncogenesis. RBL and LCL chromatin interaction maps are compared to identify the spatial and temporal genome architectural changes during EBV B cell transformation. EBV induces global genome reorganization where contact domains frequently merge or subdivide during transformation. Repressed B compartments in RBLs frequently switch to active A compartments in LCLs. LCLs gain 40% new contact domain boundaries. Newly gained LCL boundaries have strong CTCF binding at their borders while in RBLs, the same sites have much less CTCF binding. Some LCL CTCF sites also have EBV nuclear antigen (EBNA) leader protein EBNALP binding. LCLs have more local interactions than RBLs at LCL dependency factors and super-enhancer targets. RNA Pol II HiChIP and FISH of RBL and LCL further validate the Hi-C results. EBNA3A inactivation globally alters LCL genome interactions. EBNA3A inactivation reduces CTCF and RAD21 DNA binding. EBNA3C inactivation rewires the looping at the CDKN2A/B and AICDA loci. Disruption of a CTCF site at AICDA locus increases AICDA expression. These data suggest that EBV controls lymphocyte growth by globally reorganizing host genome architecture to facilitate the expression of key oncogenes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Longnecker R KE, Cohen JI. Epstein-Barr Virus, 8th edn. Lippincott Williams & Wilkins, a Wolters Kluwer Business. (2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

