Interoperable slide microscopy viewer and annotation tool for imaging data science and computational pathology
- PMID: 36949078
- PMCID: PMC10033920
- DOI: 10.1038/s41467-023-37224-2
Interoperable slide microscopy viewer and annotation tool for imaging data science and computational pathology
Abstract
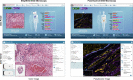
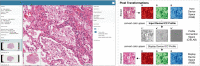
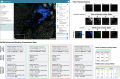
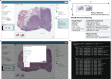
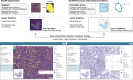
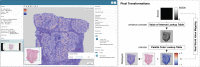
The exchange of large and complex slide microscopy imaging data in biomedical research and pathology practice is impeded by a lack of data standardization and interoperability, which is detrimental to the reproducibility of scientific findings and clinical integration of technological innovations. We introduce Slim, an open-source, web-based slide microscopy viewer that implements the internationally accepted Digital Imaging and Communications in Medicine (DICOM) standard to achieve interoperability with a multitude of existing medical imaging systems. We showcase the capabilities of Slim as the slide microscopy viewer of the NCI Imaging Data Commons and demonstrate how the viewer enables interactive visualization of traditional brightfield microscopy and highly-multiplexed immunofluorescence microscopy images from The Cancer Genome Atlas and Human Tissue Atlas Network, respectively, using standard DICOMweb services. We further show how Slim enables the collection of standardized image annotations for the development or validation of machine learning models and the visual interpretation of model inference results in the form of segmentation masks, spatial heat maps, or image-derived measurements.
© 2023. The Author(s).
Conflict of interest statement
All Authors declare the following competing interests: All Authors received funding from the National Institutes of Health for the research. David A. Clunie receives financial compensation as a consultant of Healthcare Tech Solutions (HCTS), as a consultant for Impact Business Information Solutions (IBIS), as a consultant for Mayo Foundation for Medical Education & Research, as a consultant for Essex Leidos CBIIT under under National Cancer Institute Contract No. 75N91019D00024, Task Order 75N91019F00129, as a consultant for Brigham and Women’s Hospital NCI Imaging Data Commons (IDC), as a consultant for the University of Leeds Northern Pathology Imaging Co-operative (NPIC), and as a contractor for NEMA as DICOM Editor.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

