RHOJ controls EMT-associated resistance to chemotherapy
- PMID: 36949199
- PMCID: PMC10076223
- DOI: 10.1038/s41586-023-05838-7
RHOJ controls EMT-associated resistance to chemotherapy
Abstract
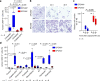
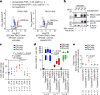
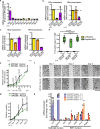
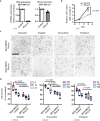
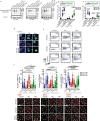
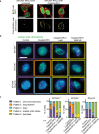
The resistance of cancer cells to therapy is responsible for the death of most patients with cancer1. Epithelial-to-mesenchymal transition (EMT) has been associated with resistance to therapy in different cancer cells2,3. However, the mechanisms by which EMT mediates resistance to therapy remain poorly understood. Here, using a mouse model of skin squamous cell carcinoma undergoing spontaneous EMT during tumorigenesis, we found that EMT tumour cells are highly resistant to a wide range of anti-cancer therapies both in vivo and in vitro. Using gain and loss of function studies in vitro and in vivo, we found that RHOJ-a small GTPase that is preferentially expressed in EMT cancer cells-controls resistance to therapy. Using genome-wide transcriptomic and proteomic profiling, we found that RHOJ regulates EMT-associated resistance to chemotherapy by enhancing the response to replicative stress and activating the DNA-damage response, enabling tumour cells to rapidly repair DNA lesions induced by chemotherapy. RHOJ interacts with proteins that regulate nuclear actin, and inhibition of actin polymerization sensitizes EMT tumour cells to chemotherapy-induced cell death in a RHOJ-dependent manner. Together, our study uncovers the role and the mechanisms through which RHOJ acts as a key regulator of EMT-associated resistance to chemotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Comment in
-
Cancer cells remodel nuclear actin filaments to resist chemotherapy.Nature. 2023 Apr;616(7955):40-42. doi: 10.1038/d41586-023-00801-y. Nature. 2023. PMID: 36949121 No abstract available.
-
EMT in chemoresistance.Nat Rev Cancer. 2023 Jun;23(6):349. doi: 10.1038/s41568-023-00581-7. Nat Rev Cancer. 2023. PMID: 37106250 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

