A CRISPR/Cas9-engineered mouse carrying a conditional knockout allele for the early growth response-1 transcription factor
- PMID: 36949241
- PMCID: PMC10514223
- DOI: 10.1002/dvg.23515
A CRISPR/Cas9-engineered mouse carrying a conditional knockout allele for the early growth response-1 transcription factor
Abstract
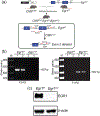
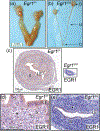
Early growth response 1 (EGR1) mediates transcriptional programs that are indispensable for cell division, differentiation, and apoptosis in numerous physiologies and pathophysiologies. Whole-body EGR1 knockouts in mice (Egr1KO ) have advanced our understanding of EGR1 function in an in vivo context. To extend the utility of the mouse to investigate EGR1 responses in a tissue- and/or cell-type-specific manner, we generated a mouse model in which exon 2 of the mouse Egr1 gene is floxed by CRISPR/Cas9 engineering. The floxed Egr1 alleles (Egr1f/f ) are designed to enable spatiotemporal control of Cre-mediated EGR1 ablation in the mouse. To confirm that the Egr1f/f alleles can be abrogated using a Cre driver, we crossed the Egr1f/f mouse with a global Cre driver to generate the Egr1 conditional knockout (Egr1d/d ) mouse in which EGR1 expression is ablated in all tissues. Genetic and protein analysis confirmed the absence of exon 2 and loss of EGR1 expression in the Egr1d/d mouse, respectively. Moreover, the Egr1d/d female exhibits overt reproductive phenotypes previously reported for the Egr1KO mouse. Therefore, studies described in this short technical report underscore the potential utility of the murine Egr1 floxed allele to further resolve EGR1 function at a tissue- and/or cell-type-specific level.
Keywords: CRISPR/Cas9; early growth response-1; floxed; mouse; ovary; pituitary; uterus.
© 2023 Wiley Periodicals LLC.
Figures






References
-
- Blaschke F, Bruemmer D, & Law RE (2004). Egr-1 is a major vascular pathogenic transcription factor in atherosclerosis and restenosis. Rev Endocr Metab Disord, 5(3), 249–254. doi:10.1023/B:REMD.0000032413.88756.ee - DOI - PubMed
-
- Davis BJ, Dixon D, & Herbert RA (1999). Ovary, oviduct, uterus, cervix, and vagina. In Maronpot RR, Boorman GA, & Gaul BW (Eds.), Pathology of the Mouse: Reference and atlas (pp. 409–444). Vienna, IL: Cache River Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

