M2 Macrophage-Derived sEV Regulate Pro-Inflammatory CCR2+ Macrophage Subpopulations to Favor Post-AMI Cardiac Repair
- PMID: 36950739
- PMCID: PMC10190454
- DOI: 10.1002/advs.202202964
M2 Macrophage-Derived sEV Regulate Pro-Inflammatory CCR2+ Macrophage Subpopulations to Favor Post-AMI Cardiac Repair
Abstract
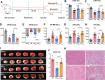
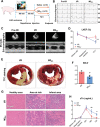
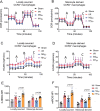
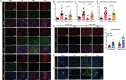
Tissue-resident cardiac macrophage subsets mediate cardiac tissue inflammation and repair after acute myocardial infarction (AMI). CC chemokine receptor 2 (CCR2)-expressing macrophages have phenotypical similarities to M1-polarized macrophages, are pro-inflammatory, and recruit CCR2+ circulating monocytes to infarcted myocardium. Small extracellular vesicles (sEV) from CCR2̶ macrophages, which phenotypically resemble M2-polarized macrophages, promote anti-inflammatory activity and cardiac repair. Here, the authors harvested M2 macrophage-derived sEV (M2EV ) from M2-polarized bone-marrow-derived macrophages for intramyocardial injection and recapitulation of sEV-mediated anti-inflammatory activity in ischemic-reperfusion (I/R) injured hearts. Rats and pigs received sham surgery; I/R without treatment; or I/R with autologous M2EV treatment. M2EV rescued cardiac function and attenuated injury markers, infarct size, and scar size. M2EV inhibited CCR2+ macrophage numbers, reduced monocyte-derived CCR2+ macrophage recruitment to infarct sites, induced M1-to-M2 macrophage switching and promoted neovascularization. Analysis of M2EV microRNA content revealed abundant miR-181b-5p, which regulated macrophage glucose uptake, glycolysis, and mitigated mitochondrial reactive oxygen species generation. Functional blockade of miR-181b-5p is detrimental to beneficial M2EV actions and resulted in failure to inhibit CCR2+ macrophage numbers and infarct size. Taken together, this investigation showed that M2EV rescued myocardial function, improved myocardial repair, and regulated CCR2+ macrophages via miR-181b-5p-dependent mechanisms, indicating an option for cell-free therapy for AMI.
Keywords: CC chemokine receptor 2; extracellular vesicles; ischemia-reperfusion injury; macrophage metabolic reprogramming; macrophages.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Reed G. W., Rossi J. E., Cannon C. P., Lancet 2017, 389, 197. - PubMed
-
- Niccoli G., Scalone G., Lerman A., Crea F., Eur. Heart J. 2016, 37, 1024. - PubMed
-
- a) Kereiakes D. J., Circulation 2003, 108, III22; - PubMed
- b) Kalkman D. N., Aquino M., Claessen B. E., Baber U., Guedeney P., Sorrentino S., Vogel B., de Winter R. J., Sweeny J., Kovacic J. C., Shah S., Vijay P., Barman N., Kini A., Sharma S., Dangas G. D., Mehran R., Eur. Heart J. 2018, 39, 4101. - PubMed
-
- Boden W. E., O'Rourke R. A., Teo K. K., Hartigan P. M., Maron D. J., Kostuk W. J., Knudtson M., Dada M., Casperson P., Harris C. L., Chaitman B. R., Shaw L., Gosselin G., Nawaz S., Title L. M., Gau G., Blaustein A. S., Booth D. C., Bates E. R., Spertus J. A., Berman D. S., Mancini G. B., Weintraub W. S., N. Engl. J. Med. 2007, 356, 1503. - PubMed
-
- a) Wang Z., Lu Y. L., Zhao W. T., Zhong J., Lin X., Sun Z., He Y., Chen M., Zheng L. R., Basic Res. Cardiol. 2020, 115, 8; - PubMed
- b) Dick S. A., Macklin J. A., Nejat S., Momen A., Clemente‐Casares X., Althagafi M. G., Chen J., Kantores C., Hosseinzadeh S., Aronoff L., Wong A., Zaman R., Barbu I., Besla R., Lavine K. J., Razani B., Ginhoux F., Husain M., Cybulsky M. I., Robbins C. S., Epelman S., Nat. Immunol. 2019, 20, 29. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
