Unmasking the tissue-resident eukaryotic DNA virome in humans
- PMID: 36951096
- PMCID: PMC10123123
- DOI: 10.1093/nar/gkad199
Unmasking the tissue-resident eukaryotic DNA virome in humans
Abstract
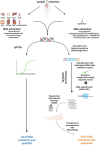
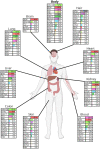
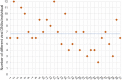
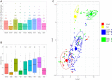
Little is known on the landscape of viruses that reside within our cells, nor on the interplay with the host imperative for their persistence. Yet, a lifetime of interactions conceivably have an imprint on our physiology and immune phenotype. In this work, we revealed the genetic make-up and unique composition of the known eukaryotic human DNA virome in nine organs (colon, liver, lung, heart, brain, kidney, skin, blood, hair) of 31 Finnish individuals. By integration of quantitative (qPCR) and qualitative (hybrid-capture sequencing) analysis, we identified the DNAs of 17 species, primarily herpes-, parvo-, papilloma- and anello-viruses (>80% prevalence), typically persisting in low copies (mean 540 copies/ million cells). We assembled in total 70 viral genomes (>90% breadth coverage), distinct in each of the individuals, and identified high sequence homology across the organs. Moreover, we detected variations in virome composition in two individuals with underlying malignant conditions. Our findings reveal unprecedented prevalences of viral DNAs in human organs and provide a fundamental ground for the investigation of disease correlates. Our results from post-mortem tissues call for investigation of the crosstalk between human DNA viruses, the host, and other microbes, as it predictably has a significant impact on our health.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I.. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023. - PubMed
-
- Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C.et al.. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015; 18:489–500. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

