Structural basis of tRNAPro acceptor stem recognition by a bacterial trans-editing domain
- PMID: 36951109
- PMCID: PMC10164551
- DOI: 10.1093/nar/gkad192
Structural basis of tRNAPro acceptor stem recognition by a bacterial trans-editing domain
Abstract
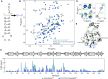
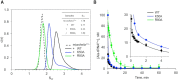
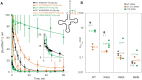
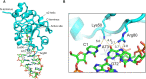
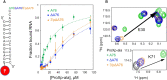
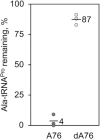
High fidelity tRNA aminoacylation by aminoacyl-tRNA synthetases is essential for cell viability. ProXp-ala is a trans-editing protein that is present in all three domains of life and is responsible for hydrolyzing mischarged Ala-tRNAPro and preventing mistranslation of proline codons. Previous studies have shown that, like bacterial prolyl-tRNA synthetase, Caulobacter crescentus ProXp-ala recognizes the unique C1:G72 terminal base pair of the tRNAPro acceptor stem, helping to ensure deacylation of Ala-tRNAPro but not Ala-tRNAAla. The structural basis for C1:G72 recognition by ProXp-ala is still unknown and was investigated here. NMR spectroscopy, binding, and activity assays revealed two conserved residues, K50 and R80, that likely interact with the first base pair, stabilizing the initial protein-RNA encounter complex. Modeling studies are consistent with direct interaction between R80 and the major groove of G72. A third key contact between A76 of tRNAPro and K45 of ProXp-ala was essential for binding and accommodating the CCA-3' end in the active site. We also demonstrated the essential role that the 2'OH of A76 plays in catalysis. Eukaryotic ProXp-ala proteins recognize the same acceptor stem positions as their bacterial counterparts, albeit with different nucleotide base identities. ProXp-ala is encoded in some human pathogens; thus, these results have the potential to inform new antibiotic drug design.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Conformational and chemical selection by a trans-acting editing domain.Proc Natl Acad Sci U S A. 2017 Aug 15;114(33):E6774-E6783. doi: 10.1073/pnas.1703925114. Epub 2017 Aug 2. Proc Natl Acad Sci U S A. 2017. PMID: 28768811 Free PMC article.
-
NMR-based solution structure of the Caulobacter crescentus ProXp-ala trans-editing enzyme.Biomol NMR Assign. 2024 Dec;18(2):233-238. doi: 10.1007/s12104-024-10193-3. Epub 2024 Aug 31. Biomol NMR Assign. 2024. PMID: 39214936 Free PMC article.
-
Human trans-editing enzyme displays tRNA acceptor-stem specificity and relaxed amino acid selectivity.J Biol Chem. 2020 Nov 27;295(48):16180-16190. doi: 10.1074/jbc.RA120.015981. Epub 2020 Oct 13. J Biol Chem. 2020. PMID: 33051185 Free PMC article.
-
Trans-editing by aminoacyl-tRNA synthetase-like editing domains.Enzymes. 2020;48:69-115. doi: 10.1016/bs.enz.2020.07.002. Epub 2020 Sep 8. Enzymes. 2020. PMID: 33837712 Review.
-
Domain-domain communication in aminoacyl-tRNA synthetases.Prog Nucleic Acid Res Mol Biol. 2001;69:317-49. doi: 10.1016/s0079-6603(01)69050-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550797 Review.
Cited by
-
Strategies for detecting aminoacylation and aminoacyl-tRNA editing in vitro and in cells.Isr J Chem. 2024 Sep;64(8-9):e202400009. doi: 10.1002/ijch.202400009. Epub 2024 May 6. Isr J Chem. 2024. PMID: 40066018 Free PMC article.
-
The role of tRNA identity elements in aminoacyl-tRNA editing.Front Microbiol. 2024 Jul 18;15:1437528. doi: 10.3389/fmicb.2024.1437528. eCollection 2024. Front Microbiol. 2024. PMID: 39101037 Free PMC article. Review.
-
Toxic Small Alarmone Synthetase FaRel2 inhibits translation by pyrophosphorylating tRNAGly and tRNAThr.bioRxiv [Preprint]. 2024 Jul 5:2024.07.05.602228. doi: 10.1101/2024.07.05.602228. bioRxiv. 2024. Update in: Sci Adv. 2024 Nov 15;10(46):eadr9624. doi: 10.1126/sciadv.adr9624. PMID: 39005314 Free PMC article. Updated. Preprint.
-
Toxic small alarmone synthetase FaRel2 inhibits translation by pyrophosphorylating tRNAGly and tRNAThr.Sci Adv. 2024 Nov 15;10(46):eadr9624. doi: 10.1126/sciadv.adr9624. Epub 2024 Nov 13. Sci Adv. 2024. PMID: 39536105 Free PMC article.
-
Mistranslating the genetic code with leucine in yeast and mammalian cells.RNA Biol. 2024 Jan;21(1):1-23. doi: 10.1080/15476286.2024.2340297. Epub 2024 Apr 17. RNA Biol. 2024. PMID: 38629491 Free PMC article.
References
-
- Ling J., Reynolds N., Ibba M.. Aminoacyl-tRNA synthesis and translational Quality control. Annu. Rev. Microbiol. 2009; 63:61–78. - PubMed
-
- Mascarenhas A.P., An S., Rosen A.E., Martinis S.A.. Köhler C., Rajbhandary U.L.. Fidelity mechanisms of the aminoacyl-tRNA synthetases. Protein Engineering. 2008; NY: Springer; 153–200.
-
- Yadavalli B.S.S., Ibba M.. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv. Protein Chem. Struct. Biol. 2012; 86:1–43. - PubMed
-
- Lee J.W., Beebe K., Nangle L.A., Jang J., Longo-Guess C.M., Cook S.A., Davisson M.T., Sundberg J.P., Schimmel P., Ackerman S.L.. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006; 443:50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

