Enterohemorrhagic Escherichia coli responds to gut microbiota metabolites by altering metabolism and activating stress responses
- PMID: 36951510
- PMCID: PMC10038027
- DOI: 10.1080/19490976.2023.2190303
Enterohemorrhagic Escherichia coli responds to gut microbiota metabolites by altering metabolism and activating stress responses
Abstract
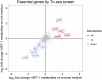
Enterohemorrhagic Escherichia coli (EHEC) is a major cause of severe bloody diarrhea, with potentially lethal complications, such as hemolytic uremic syndrome. In humans, EHEC colonizes the colon, which is also home to a diverse community of trillions of microbes known as the gut microbiota. Although these microbes and the metabolites that they produce represent an important component of EHEC's ecological niche, little is known about how EHEC senses and responds to the presence of gut microbiota metabolites. In this study, we used a combined RNA-Seq and Tn-Seq approach to characterize EHEC's response to metabolites from an in vitro culture of 33 human gut microbiota isolates (MET-1), previously demonstrated to effectively resolve recurrent Clostridioides difficile infection in human patients. Collectively, the results revealed that EHEC adjusts to growth in the presence of microbiota metabolites in two major ways: by altering its metabolism and by activating stress responses. Metabolic adaptations to the presence of microbiota metabolites included increased expression of systems for maintaining redox balance and decreased expression of biotin biosynthesis genes, reflecting the high levels of biotin released by the microbiota into the culture medium. In addition, numerous genes related to envelope and oxidative stress responses (including cpxP, spy, soxS, yhcN, and bhsA) were upregulated during EHEC growth in a medium containing microbiota metabolites. Together, these results provide insight into the molecular mechanisms by which pathogens adapt to the presence of competing microbes in the host environment, which ultimately may enable the development of therapies to enhance colonization resistance and prevent infection.
Keywords: EHEC; Enterohemorrhagic Escherichia coli; RNA-seq; Tn-seq; bacterial metabolism; gut microbiota; metabolites; stress response.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


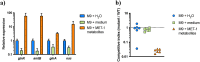
Similar articles
-
Interactions between Enterohemorrhagic Escherichia coli (EHEC) and Gut Commensals at the Interface of Human Colonoids.mBio. 2022 Jun 28;13(3):e0132122. doi: 10.1128/mbio.01321-22. Epub 2022 May 31. mBio. 2022. PMID: 35638758 Free PMC article.
-
Impact of Western Diet on Enterohemorrhagic Escherichia coli Colonization in the Human In Vitro Mucosal Artificial Colon as Mediated by Gut Microbiota.Nutrients. 2024 Jun 27;16(13):2046. doi: 10.3390/nu16132046. Nutrients. 2024. PMID: 38999794 Free PMC article.
-
Catabolite and Oxygen Regulation of Enterohemorrhagic Escherichia coli Virulence.mBio. 2016 Nov 22;7(6):e01852-16. doi: 10.1128/mBio.01852-16. mBio. 2016. PMID: 27879335 Free PMC article.
-
Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract.Virulence. 2013 May 15;4(4):315-23. doi: 10.4161/viru.24318. Epub 2013 Apr 3. Virulence. 2013. PMID: 23552827 Free PMC article. Review.
-
The Interplay between the Microbiota and Enterohemorrhagic Escherichia coli.Microbiol Spectr. 2014 Oct;2(5). doi: 10.1128/microbiolspec.EHEC-0015-2013. Microbiol Spectr. 2014. PMID: 26104352 Review.
Cited by
-
Throwing a spotlight on genomic dark matter: The power and potential of transposon-insertion sequencing.J Biol Chem. 2025 Jun;301(6):110231. doi: 10.1016/j.jbc.2025.110231. Epub 2025 May 14. J Biol Chem. 2025. PMID: 40378959 Free PMC article. Review.
-
Role of dietary nutrients and metabolism in colorectal cancer.Asia Pac J Clin Nutr. 2024 Jun;33(2):153-161. doi: 10.6133/apjcn.202406_33(2).0002. Asia Pac J Clin Nutr. 2024. PMID: 38794975 Free PMC article. Review.
-
Fitness gene Characterization of Human Infant-Derived Commensal Escherichia coli LFYP5 in the Murine Gut.Curr Microbiol. 2025 Jul 14;82(9):383. doi: 10.1007/s00284-025-04358-z. Curr Microbiol. 2025. PMID: 40658262
-
Biotin Homeostasis and Human Disorders: Recent Findings and Perspectives.Int J Mol Sci. 2024 Jun 14;25(12):6578. doi: 10.3390/ijms25126578. Int J Mol Sci. 2024. PMID: 38928282 Free PMC article. Review.
References
-
- Fatima R, Aziz M.. Enterohemorrhagic Escherichia coli. StatPearls. 2022. PMID: 30137794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
