Polydopamine Nanoparticles Targeting Ferroptosis Mitigate Intervertebral Disc Degeneration Via Reactive Oxygen Species Depletion, Iron Ions Chelation, and GPX4 Ubiquitination Suppression
- PMID: 36951540
- PMCID: PMC10161035
- DOI: 10.1002/advs.202207216
Polydopamine Nanoparticles Targeting Ferroptosis Mitigate Intervertebral Disc Degeneration Via Reactive Oxygen Species Depletion, Iron Ions Chelation, and GPX4 Ubiquitination Suppression
Abstract
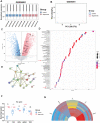
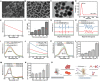
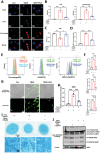
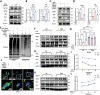
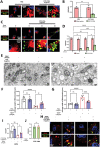
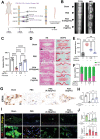
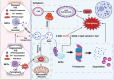
Intervertebral disc degeneration (IVDD)-induced lower back pain (LBP) is a common problem worldwide. The underlying mechanism is partially accredited to ferroptosis, based on sequencing analyses of IVDD patients from the gene expression omnibus (GEO) databases. In this study, it is shown that polydopamine nanoparticles (PDA NPs) inhibit oxidative stress-induced ferroptosis in nucleus pulposus (NP) cells in vitro. PDA NPs scavenge reactive oxygen species (ROS), chelate Fe2+ to mitigate iron overload, and regulate the expression of iron storage proteins such as ferritin heavy chain (FHC), ferritin, and transferrin receptor (TFR). More importantly, PDA NPs co-localize with glutathione peroxidase 4 (GPX4) around the mitochondria and suppress ubiquitin-mediated degradation, which in turn exerts a protective function via the transformation and clearance of phospholipid hydroperoxides. PDA NPs further down-regulate malondialdehyde (MDA) and lipid peroxide (LPO) production; thus, antagonizing ferroptosis in NP cells. Moreover, PDA NPs effectively rescue puncture-induced degeneration in vivo by targeting ferroptosis and inhibiting GPX4 ubiquitination, resulting in the upregulation of antioxidant pathways. The findings offer a new tool to explore the underlying mechanisms and a novel treatment strategy for IVDD-induced LBP.
Keywords: GPX4 ubiquitination; ferroptosis; intervertebral disc degeneration; nucleus pulposus; polydopamine nanoparticles.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Nordihydroguaiaretic acid suppresses ferroptosis and mitigates intervertebral disc degeneration through the NRF2/GPX4 axis.Int Immunopharmacol. 2024 Dec 25;143(Pt 3):113590. doi: 10.1016/j.intimp.2024.113590. Epub 2024 Nov 15. Int Immunopharmacol. 2024. PMID: 39541847
-
Ferroptosis: A New Direction in the Treatment of Intervertebral Disc Degeneration.Cell Biochem Biophys. 2025 Mar;83(1):33-42. doi: 10.1007/s12013-024-01468-6. Epub 2024 Aug 5. Cell Biochem Biophys. 2025. PMID: 39102089 Review.
-
Selenium-SelK-GPX4 axis protects nucleus pulposus cells against mechanical overloading-induced ferroptosis and attenuates senescence of intervertebral disc.Cell Mol Life Sci. 2024 Jan 22;81(1):49. doi: 10.1007/s00018-023-05067-1. Cell Mol Life Sci. 2024. PMID: 38252317 Free PMC article.
-
Cynarin alleviates intervertebral disc degeneration via protecting nucleus pulposus cells from ferroptosis.Biomed Pharmacother. 2023 Sep;165:115252. doi: 10.1016/j.biopha.2023.115252. Epub 2023 Aug 1. Biomed Pharmacother. 2023. PMID: 37536034
-
Ferroptosis: A potential target for the intervention of intervertebral disc degeneration.Front Endocrinol (Lausanne). 2022 Oct 20;13:1042060. doi: 10.3389/fendo.2022.1042060. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36339421 Free PMC article. Review.
Cited by
-
Application trends and strategies of hydrogel delivery systems in intervertebral disc degeneration: A bibliometric review.Mater Today Bio. 2024 Sep 14;28:101251. doi: 10.1016/j.mtbio.2024.101251. eCollection 2024 Oct. Mater Today Bio. 2024. PMID: 39318370 Free PMC article. Review.
-
Role of non‑coding RNAs in cartilage endplate (Review).Exp Ther Med. 2023 May 11;26(1):312. doi: 10.3892/etm.2023.12011. eCollection 2023 Jul. Exp Ther Med. 2023. PMID: 37273754 Free PMC article. Review.
-
Iron Overload in Brain: Transport Mismatches, Microbleeding Events, and How Nanochelating Therapies May Counteract Their Effects.Int J Mol Sci. 2024 Feb 16;25(4):2337. doi: 10.3390/ijms25042337. Int J Mol Sci. 2024. PMID: 38397013 Free PMC article. Review.
-
The Role and Interactive Mechanism of Endoplasmic Reticulum Stress and Ferroptosis in Musculoskeletal Disorders.Biomolecules. 2024 Oct 28;14(11):1369. doi: 10.3390/biom14111369. Biomolecules. 2024. PMID: 39595546 Free PMC article. Review.
-
The TF/Nrf2/GSTP1 pathway is involved in stress-induced hepatocellular injury through ferroptosis.J Cell Mol Med. 2024 Jun;28(12):e18494. doi: 10.1111/jcmm.18494. J Cell Mol Med. 2024. PMID: 38890797 Free PMC article.
References
-
- Hartvigsen J., Hancock M. J., Kongsted A., Louw Q., Ferreira M. L., Genevay S., Hoy D., Karppinen J., Pransky G., Sieper J., Smeets R. J., Underwood M., Lancet 2018, 391, 2356. - PubMed
-
- DePalma M. J., Ketchum J. M., Saullo T., Pain Medicine 2011, 12, 224. - PubMed
-
- Chou D., Samartzis D., Bellabarba C., Patel A., Luk K. D. K., Kisser J. M. S., Skelly A. C., Spine 2011, 36, S43. - PubMed
-
- Takatalo J., Karppinen J., Niinimäki J., Taimela S., Näyhä S., Mutanen P., Sequeiros R. B., Kyllönen E., Tervonen O., Spine 2011, 36, 2180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
