Bacteria in Fluid Flow
- PMID: 36951552
- PMCID: PMC10127660
- DOI: 10.1128/jb.00400-22
Bacteria in Fluid Flow
Abstract
Bacteria thrive in environments rich in fluid flow, such as the gastrointestinal tract, bloodstream, aquatic systems, and the urinary tract. Despite the importance of flow, how flow affects bacterial life is underappreciated. In recent years, the combination of approaches from biology, physics, and engineering has led to a deeper understanding of how bacteria interact with flow. Here, we highlight the wide range of bacterial responses to flow, including changes in surface adhesion, motility, surface colonization, quorum sensing, virulence factor production, and gene expression. To emphasize the diversity of flow responses, we focus our review on how flow affects four ecologically distinct bacterial species: Escherichia coli, Staphylococcus aureus, Caulobacter crescentus, and Pseudomonas aeruginosa. Additionally, we present experimental approaches to precisely study bacteria in flow, discuss how only some flow responses are triggered by shear force, and provide perspective on flow-sensitive bacterial signaling.
Keywords: adhesion; bacteria; colonization; fluid flow; gene expression; mechanobiology; mechanosensing; motility; quorum sensing; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
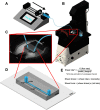
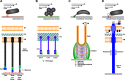

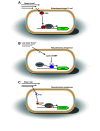
Similar articles
-
Subinhibitory concentrations of phenyl lactic acid interfere with the expression of virulence factors in Staphylococcus aureus and Pseudomonas aeruginosa clinical strains.Roum Arch Microbiol Immunol. 2009 Jan-Mar;68(1):27-33. Roum Arch Microbiol Immunol. 2009. PMID: 19507624
-
Bacterial quorum sensing: its role in virulence and possibilities for its control.Cold Spring Harb Perspect Med. 2012 Nov 1;2(11):a012427. doi: 10.1101/cshperspect.a012427. Cold Spring Harb Perspect Med. 2012. PMID: 23125205 Free PMC article. Review.
-
Anti-quorum Sensing and Anti-biofilm Activity of Delftia tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa.Front Cell Infect Microbiol. 2017 Jul 26;7:337. doi: 10.3389/fcimb.2017.00337. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28798903 Free PMC article.
-
Identification of a novel regulator of the quorum-sensing systems in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2009 Apr;293(2):196-204. doi: 10.1111/j.1574-6968.2009.01544.x. Epub 2009 Mar 2. FEMS Microbiol Lett. 2009. PMID: 19260962
-
Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction.Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):712-728. doi: 10.1080/1040841X.2019.1700209. Epub 2019 Dec 13. Crit Rev Microbiol. 2019. PMID: 31835971 Review.
Cited by
-
Effect of host microenvironment and bacterial lifestyles on antimicrobial sensitivity and implications for susceptibility testing.NPJ Antimicrob Resist. 2025 May 21;3(1):42. doi: 10.1038/s44259-025-00113-3. NPJ Antimicrob Resist. 2025. PMID: 40399473 Free PMC article. Review.
-
Shear force enhances adhesion of Pseudomonas aeruginosa by counteracting pilus-driven surface departure.Proc Natl Acad Sci U S A. 2023 Oct 10;120(41):e2307718120. doi: 10.1073/pnas.2307718120. Epub 2023 Oct 3. Proc Natl Acad Sci U S A. 2023. PMID: 37788310 Free PMC article.
-
Flagellum-driven motility enhances Pseudomonas aeruginosa biofilm formation by altering cell orientation.Appl Environ Microbiol. 2025 Jul 23;91(7):e0082125. doi: 10.1128/aem.00821-25. Epub 2025 Jul 3. Appl Environ Microbiol. 2025. PMID: 40607807 Free PMC article.
-
The Impact of Fluid Flow on Microbial Growth and Distribution in Food Processing Systems.Foods. 2025 Jan 26;14(3):401. doi: 10.3390/foods14030401. Foods. 2025. PMID: 39941998 Free PMC article. Review.
-
Effect of Gravity on Bacterial Adhesion to Heterogeneous Surfaces.Pathogens. 2023 Jul 15;12(7):941. doi: 10.3390/pathogens12070941. Pathogens. 2023. PMID: 37513788 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

