Impact of the Dimethyl Sulfoxide Reductase Superfamily on the Evolution of Biogeochemical Cycles
- PMID: 36951557
- PMCID: PMC10100899
- DOI: 10.1128/spectrum.04145-22
Impact of the Dimethyl Sulfoxide Reductase Superfamily on the Evolution of Biogeochemical Cycles
Abstract
The dimethyl sulfoxide reductase (or MopB) family is a diverse assemblage of enzymes found throughout Bacteria and Archaea. Many of these enzymes are believed to have been present in the last universal common ancestor (LUCA) of all cellular lineages. However, gaps in knowledge remain about how MopB enzymes evolved and how this diversification of functions impacted global biogeochemical cycles through geologic time. In this study, we perform maximum likelihood phylogenetic analyses on manually curated comparative genomic and metagenomic data sets containing over 47,000 distinct MopB homologs. We demonstrate that these enzymes constitute a catalytically and mechanistically diverse superfamily defined not by the molybdopterin- or tungstopterin-containing [molybdopterin or tungstopterin bis(pyranopterin guanine dinucleotide) (Mo/W-bisPGD)] cofactor but rather by the structural fold that binds it in the protein. Our results suggest that major metabolic innovations were the result of the loss of the metal cofactor or the gain or loss of protein domains. Phylogenetic analyses also demonstrated that formate oxidation and CO2 reduction were the ancestral functions of the superfamily, traits that have been vertically inherited from the LUCA. Nearly all of the other families, which drive all other biogeochemical cycles mediated by this superfamily, originated in the bacterial domain. Thus, organisms from Bacteria have been the key drivers of catalytic and biogeochemical innovations within the superfamily. The relative ordination of MopB families and their associated catalytic activities emphasize fundamental mechanisms of evolution in this superfamily. Furthermore, it underscores the importance of prokaryotic adaptability in response to the transition from an anoxic to an oxidized atmosphere. IMPORTANCE The MopB superfamily constitutes a repertoire of metalloenzymes that are central to enduring mysteries in microbiology, from the origin of life and how microorganisms and biogeochemical cycles have coevolved over deep time to how anaerobic life adapted to increasing concentrations of O2 during the transition from an anoxic to an oxic world. Our work emphasizes that phylogenetic analyses can reveal how domain gain or loss events, the acquisition of novel partner subunits, and the loss of metal cofactors can stimulate novel radiations of enzymes that dramatically increase the catalytic versatility of superfamilies. We also contend that the superfamily concept in protein evolution can uncover surprising kinships between enzymes that have remarkably different catalytic and physiological functions.
Keywords: DMSO reductase; MopB; biogeochemical cycles; evolution; molybdopterin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


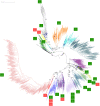
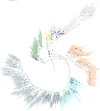

References
LinkOut - more resources
Full Text Sources
Miscellaneous

