Stromal and therapy-induced macrophage proliferation promotes PDAC progression and susceptibility to innate immunotherapy
- PMID: 36951731
- PMCID: PMC10072222
- DOI: 10.1084/jem.20212062
Stromal and therapy-induced macrophage proliferation promotes PDAC progression and susceptibility to innate immunotherapy
Abstract
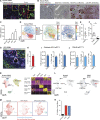
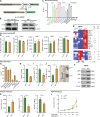
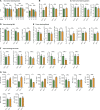
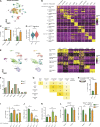
Tumor-associated macrophages (TAMs) are abundant in pancreatic ductal adenocarcinomas (PDACs). While TAMs are known to proliferate in cancer tissues, the impact of this on macrophage phenotype and disease progression is poorly understood. We showed that in PDAC, proliferation of TAMs could be driven by colony stimulating factor-1 (CSF1) produced by cancer-associated fibroblasts. CSF1 induced high levels of p21 in macrophages, which regulated both TAM proliferation and phenotype. TAMs in human and mouse PDACs with high levels of p21 had more inflammatory and immunosuppressive phenotypes. p21 expression in TAMs was induced by both stromal interaction and/or chemotherapy treatment. Finally, by modeling p21 expression levels in TAMs, we found that p21-driven macrophage immunosuppression in vivo drove tumor progression. Serendipitously, the same p21-driven pathways that drive tumor progression also drove response to CD40 agonist. These data suggest that stromal or therapy-induced regulation of cell cycle machinery can regulate both macrophage-mediated immune suppression and susceptibility to innate immunotherapy.
© 2023 Zuo et al.
Conflict of interest statement
Disclosures: J.M. Baer reported grants from the National Institutes of Health during the conduct of the study. No other disclosures were reported.
Figures









References
-
- Borgoni, S., Iannello A., Cutrupi S., Allavena P., D’Incalci M., Novelli F., and Cappello P.. 2017. Depletion of tumor-associated macrophages switches the epigenetic profile of pancreatic cancer infiltrating T cells and restores their anti-tumor phenotype. OncoImmunology. 7:e1393596. 10.1080/2162402X.2017.1393596 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

