Intrathecal AAV9/AP4M1 gene therapy for hereditary spastic paraplegia 50 shows safety and efficacy in preclinical studies
- PMID: 36951961
- PMCID: PMC10178841
- DOI: 10.1172/JCI164575
Intrathecal AAV9/AP4M1 gene therapy for hereditary spastic paraplegia 50 shows safety and efficacy in preclinical studies
Abstract
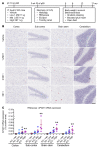
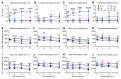
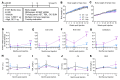
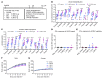
Spastic paraplegia 50 (SPG50) is an ultrarare childhood-onset neurological disorder caused by biallelic loss-of-function variants in the AP4M1 gene. SPG50 is characterized by progressive spastic paraplegia, global developmental delay, and subsequent intellectual disability, secondary microcephaly, and epilepsy. We preformed preclinical studies evaluating an adeno-associated virus (AAV)/AP4M1 gene therapy for SPG50 and describe in vitro studies that demonstrate transduction of patient-derived fibroblasts with AAV2/AP4M1, resulting in phenotypic rescue. To evaluate efficacy in vivo, Ap4m1-KO mice were intrathecally (i.t.) injected with 5 × 1011, 2.5 × 1011, or 1.25 × 1011 vector genome (vg) doses of AAV9/AP4M1 at P7-P10 or P90. Age- and dose-dependent effects were observed, with early intervention and higher doses achieving the best therapeutic benefits. In parallel, three toxicology studies in WT mice, rats, and nonhuman primates (NHPs) demonstrated that AAV9/AP4M1 had an acceptable safety profile up to a target human dose of 1 × 1015 vg. Of note, similar degrees of minimal-to-mild dorsal root ganglia (DRG) toxicity were observed in both rats and NHPs, supporting the use of rats to monitor DRG toxicity in future i.t. AAV studies. These preclinical results identify an acceptably safe and efficacious dose of i.t.-administered AAV9/AP4M1, supporting an investigational gene transfer clinical trial to treat SPG50.
Keywords: Neurological disorders; Neuroscience.
Conflict of interest statement
Figures




Comment in
-
Paving a way to treat spastic paraplegia 50.J Clin Invest. 2023 May 15;133(10):e170226. doi: 10.1172/JCI170226. J Clin Invest. 2023. PMID: 37183815 Free PMC article.
References
-
- Ebrahimi-Fakhari D, et al. AP-4-Associated hereditary spastic paraplegia. In: Adam MP, et al., eds. GeneReviews. University of Washington; 2018:1993–2023. https://www.ncbi.nlm.nih.gov/books/NBK535153/ Accessed May 3, 2023.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

