Antigen discrimination by T cells relies on size-constrained microvillar contact
- PMID: 36959206
- PMCID: PMC10036606
- DOI: 10.1038/s41467-023-36855-9
Antigen discrimination by T cells relies on size-constrained microvillar contact
Abstract
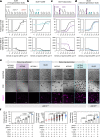
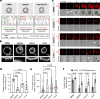
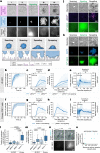
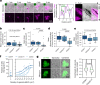
T cells use finger-like protrusions called 'microvilli' to interrogate their targets, but why they do so is unknown. To form contacts, T cells must overcome the highly charged, barrier-like layer of large molecules forming a target cell's glycocalyx. Here, T cells are observed to use microvilli to breach a model glycocalyx barrier, forming numerous small (<0.5 μm diameter) contacts each of which is stabilized by the small adhesive protein CD2 expressed by the T cell, and excludes large proteins including CD45, allowing sensitive, antigen dependent TCR signaling. In the absence of the glycocalyx or when microvillar contact-size is increased by enhancing CD2 expression, strong signaling occurs that is no longer antigen dependent. Our observations suggest that, modulated by the opposing effects of the target cell glycocalyx and small adhesive proteins, the use of microvilli equips T cells with the ability to effect discriminatory receptor signaling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

