ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages
- PMID: 36959508
- PMCID: PMC10159855
- DOI: 10.1038/s41564-023-01335-9
ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages
Abstract
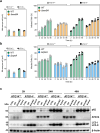
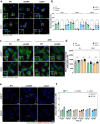
Autophagy is a cellular innate-immune defence mechanism against intracellular microorganisms, including Mycobacterium tuberculosis (Mtb). How canonical and non-canonical autophagy function to control Mtb infection in phagosomes and the cytosol remains unresolved. Macrophages are the main host cell in humans for Mtb. Here we studied the contributions of canonical and non-canonical autophagy in the genetically tractable human induced pluripotent stem cell-derived macrophages (iPSDM), using a set of Mtb mutants generated in the same genetic background of the common lab strain H37Rv. We monitored replication of Mtb mutants that are either unable to trigger canonical autophagy (Mtb ΔesxBA) or reportedly unable to block non-canonical autophagy (Mtb ΔcpsA) in iPSDM lacking either ATG7 or ATG14 using single-cell high-content imaging. We report that deletion of ATG7 by CRISPR-Cas9 in iPSDM resulted in increased replication of wild-type Mtb but not of Mtb ΔesxBA or Mtb ΔcpsA. We show that deletion of ATG14 resulted in increased replication of both Mtb wild type and the mutant Mtb ΔesxBA. Using Mtb reporters and quantitative imaging, we identified a role for ATG14 in regulating fusion of phagosomes containing Mtb with lysosomes, thereby enabling intracellular bacteria restriction. We conclude that ATG7 and ATG14 are both required for restricting Mtb replication in human macrophages.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Comment in
-
Autophagy is part of the answer to tuberculosis.Nat Microbiol. 2023 May;8(5):762-763. doi: 10.1038/s41564-023-01373-3. Nat Microbiol. 2023. PMID: 37142685 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

