Analysis of the molecular and biochemical mechanisms involved in the symbiotic relationship between Arbuscular mycorrhiza fungi and Manihot esculenta Crantz
- PMID: 36959933
- PMCID: PMC10028151
- DOI: 10.3389/fpls.2023.1130924
Analysis of the molecular and biochemical mechanisms involved in the symbiotic relationship between Arbuscular mycorrhiza fungi and Manihot esculenta Crantz
Abstract
Introduction: Plants and arbuscular mycorrhizal fungi (AMF) mutualistic interactions are essential for sustainable agriculture production. Although it is shown that AMF inoculation improves cassava physiological performances and yield traits, the molecular mechanisms involved in AM symbiosis remain largely unknown. Herein, we integrated metabolomics and transcriptomics analyses of symbiotic (Ri) and asymbiotic (CK) cassava roots and explored AM-induced biochemical and transcriptional changes.
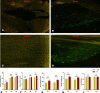
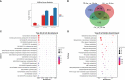
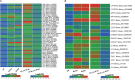
Results: Three weeks (3w) after AMF inoculations, proliferating fungal hyphae were observable, and plant height and root length were significantly increased. In total, we identified 1,016 metabolites, of which 25 were differentially accumulated (DAMs) at 3w. The most highly induced metabolites were 5-aminolevulinic acid, L-glutamic acid, and lysoPC 18:2. Transcriptome analysis identified 693 and 6,481 differentially expressed genes (DEGs) in the comparison between CK (3w) against Ri at 3w and 6w, respectively. Functional enrichment analyses of DAMs and DEGs unveiled transport, amino acids and sugar metabolisms, biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, and plant-pathogen interactions as the most differentially regulated pathways. Potential candidate genes, including nitrogen and phosphate transporters, transcription factors, phytohormone, sugar metabolism-related, and SYM (symbiosis) signaling pathway-related, were identified for future functional studies.
Discussion: Our results provide molecular insights into AM symbiosis and valuable resources for improving cassava production.
Keywords: arbuscular mycorrhiza; candidate gene; cassava; fungi; metabolome; symbiosis; transcriptome.
Copyright © 2023 Gao, Huang, Wang, Lin, Pan, Zhang, Zhang, Wang, Cheng and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Abdallah C., Valot B., Guillier C., Mounier A., Balliau T., Zivy M., et al. (2014). The membrane proteome of medicago truncatula roots displays qualitative and quantitative changes in response to arbuscular mycorrhizal symbiosis. J. Proteomics 108, 354–368. doi: 10.1016/j.jprot.2014.05.028. Elsevier B.V. - DOI - PubMed
-
- Aliyu I. A., Yusuf A. A., Uyovbisere E. O., Masso C., Sanders I. R. (2018). Effect of co-application of phosphorus fertilizer and in vitro-produced mycorrhizal fungal inoculants on yield and leaf nutrient concentration of cassava. PLos One 14, 1–19. doi: 10.1371/journal.pone.0218969 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

