Simulated-to-real benchmarking of acquisition methods in untargeted metabolomics
- PMID: 36959982
- PMCID: PMC10027714
- DOI: 10.3389/fmolb.2023.1130781
Simulated-to-real benchmarking of acquisition methods in untargeted metabolomics
Abstract
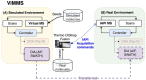
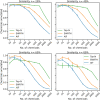
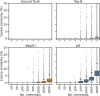
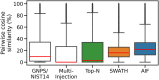
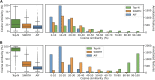
Data-Dependent and Data-Independent Acquisition modes (DDA and DIA, respectively) are both widely used to acquire MS2 spectra in untargeted liquid chromatography tandem mass spectrometry (LC-MS/MS) metabolomics analyses. Despite their wide use, little work has been attempted to systematically compare their MS/MS spectral annotation performance in untargeted settings due to the lack of ground truth and the costs involved in running a large number of acquisitions. Here, we present a systematic in silico comparison of these two acquisition methods in untargeted metabolomics by extending our Virtual Metabolomics Mass Spectrometer (ViMMS) framework with a DIA module. Our results show that the performance of these methods varies with the average number of co-eluting ions as the most important factor. At low numbers, DIA outperforms DDA, but at higher numbers, DDA has an advantage as DIA can no longer deal with the large amount of overlapping ion chromatograms. Results from simulation were further validated on an actual mass spectrometer, demonstrating that using ViMMS we can draw conclusions from simulation that translate well into the real world. The versatility of the Virtual Metabolomics Mass Spectrometer (ViMMS) framework in simulating different parameters of both Data-Dependent and Data-Independent Acquisition (DDA and DIA) modes is a key advantage of this work. Researchers can easily explore and compare the performance of different acquisition methods within the ViMMS framework, without the need for expensive and time-consuming experiments with real experimental data. By identifying the strengths and limitations of each acquisition method, researchers can optimize their choice and obtain more accurate and robust results. Furthermore, the ability to simulate and validate results using the ViMMS framework can save significant time and resources, as it eliminates the need for numerous experiments. This work not only provides valuable insights into the performance of DDA and DIA, but it also opens the door for further advancements in LC-MS/MS data acquisition methods.
Keywords: data independent acquisition; data-dependent acquisition; digital twin; liquid chromatography tandem mass spectrometry; metabolomics.
Copyright © 2023 Wandy, McBride, Rogers, Terzis, Weidt, van der Hooft, Bryson, Daly and Davies.
Conflict of interest statement
JJJVdH is member of the Scientific Advisory Board of NAICONS Srl. Milano, Italy. All other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., et al. (2012). Targeted data extraction of the ms/ms spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. 10.1074/mcp.O111.016717 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

