AlphaFold predicts novel human proteins with knots
- PMID: 36960558
- PMCID: PMC10108431
- DOI: 10.1002/pro.4631
AlphaFold predicts novel human proteins with knots
Abstract
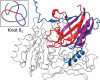
The fact that proteins can have their chain formed in a knot is known for almost 30 years. However, as they are not common, only a fraction of such proteins is available in the Protein Data Bank. It was not possible to assess their importance and versatility up until now because we did not have access to the whole proteome of an organism, let alone a human one. The arrival of efficient machine learning methods for protein structure prediction, such as AlphaFold and RoseTTaFold, changed that. We analyzed all proteins from the human proteome (over 20,000) determined with AlphaFold in search for knots and found them in less than 2% of the structures. Using a variety of methods, including homolog search, clustering, quality assessment, and visual inspection, we determined the nature of each of the knotted structures and classified it as either knotted, potentially knotted, or an artifact, and deposited all of them in a database available at: https://knotprot.cent.uw.edu.pl/alphafold. Overall, we found 51 credible knotted proteins (0.2% of human proteome). The set of potentially knotted structures includes a new complex type of a knot not reported in proteins yet. That knot type, denoted 63 in mathematical notation, would necessitate a more complex folding path than any knotted protein characterized to date.
Keywords: biological function; evolution; folding; new knotted folds.
© 2023 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Alexander JW. Topological invariants of knots and links. Trans Am Math So. 1928;30(2):275–306.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

