Whole Genome Analysis of Venous Thromboembolism: the Trans-Omics for Precision Medicine Program
- PMID: 36960714
- PMCID: PMC10151032
- DOI: 10.1161/CIRCGEN.121.003532
Whole Genome Analysis of Venous Thromboembolism: the Trans-Omics for Precision Medicine Program
Abstract
Background: Risk for venous thromboembolism has a strong genetic component. Whole genome sequencing from the TOPMed program (Trans-Omics for Precision Medicine) allowed us to look for new associations, particularly rare variants missed by standard genome-wide association studies.
Methods: The 3793 cases and 7834 controls (11.6% of cases were individuals of African, Hispanic/Latino, or Asian ancestry) were analyzed using a single variant approach and an aggregate gene-based approach using our primary filter (included only loss-of-function and missense variants predicted to be deleterious) and our secondary filter (included all missense variants).
Results: Single variant analyses identified associations at 5 known loci. Aggregate gene-based analyses identified only PROC (odds ratio, 6.2 for carriers of rare variants; P=7.4×10-14) when using our primary filter. Employing our secondary variant filter led to a smaller effect size at PROC (odds ratio, 3.8; P=1.6×10-14), while excluding variants found only in rare isoforms led to a larger one (odds ratio, 7.5). Different filtering strategies improved the signal for 2 other known genes: PROS1 became significant (minimum P=1.8×10-6 with the secondary filter), while SERPINC1 did not (minimum P=4.4×10-5 with minor allele frequency <0.0005). Results were largely the same when restricting the analyses to include only unprovoked cases; however, one novel gene, MS4A1, became significant (P=4.4×10-7 using all missense variants with minor allele frequency <0.0005).
Conclusions: Here, we have demonstrated the importance of using multiple variant filtering strategies, as we detected additional genes when filtering variants based on their predicted deleteriousness, frequency, and presence on the most expressed isoforms. Our primary analyses did not identify new candidate loci; thus larger follow-up studies are needed to replicate the novel MS4A1 locus and to identify additional rare variation associated with venous thromboembolism.
Keywords: cardiovascular diseases; genetics; genome-wide association study; single nucleotide polymorphisms; venous thromboembolism; whole genome sequencing.
Figures
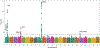
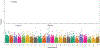
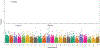
References
-
- Silverstein MD, Heit JA, Mohr DN, Petterson TM, O’Fallon WM, Melton LJ 3rd. Trends in the incidence of deep vein thrombosis and pulmonary embolism: a 25-year population-based study. Arch Intern Med. 1998;158:585–593. - PubMed
-
- Wolberg AS, Rosendaal FR, Weitz JI, Jaffer IH, Agnelli G, Baglin T, Mackman N. Venous thrombosis. Nat Rev Dis Primers. 2015;1:15006. - PubMed
-
- Heit JA, Phelps MA, Ward SA, Slusser JP, Petterson TM, De Andrade M. Familial segregation of venous thromboembolism. J Thromb Haemost. 2004;2:731–736. - PubMed
-
- Larsen TB, Sørensen HT, Skytthe A, Johnsen SP, Vaupel JW, Christensen K. Major genetic susceptibility for venous thromboembolism in men: a study of Danish twins. Epidemiology. 2003;14:328–332. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

