Clinical PARP inhibitors allosterically induce PARP2 retention on DNA
- PMID: 36961901
- PMCID: PMC10038340
- DOI: 10.1126/sciadv.adf7175
Clinical PARP inhibitors allosterically induce PARP2 retention on DNA
Abstract
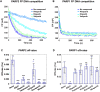
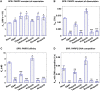
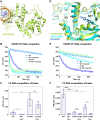
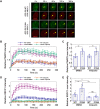
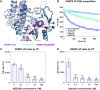
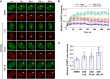
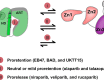
PARP1 and PARP2 detect DNA breaks, which activates their catalytic production of poly(ADP-ribose) that recruits repair factors and contributes to PARP1/2 release from DNA. PARP inhibitors (PARPi) are used in cancer treatment and target PARP1/2 catalytic activity, interfering with repair and increasing PARP1/2 persistence on DNA damage. In addition, certain PARPi exert allosteric effects that increase PARP1 retention on DNA. However, no clinical PARPi exhibit this allosteric behavior toward PARP1. In contrast, we show that certain clinical PARPi exhibit an allosteric effect that retains PARP2 on DNA breaks in a manner that depends on communication between the catalytic and DNA binding regions. Using a PARP2 mutant that mimics an allosteric inhibitor effect, we observed increased PARP2 retention at cellular damage sites. The PARPi AZD5305 also exhibited a clear reverse allosteric effect on PARP2. Our results can help explain the toxicity of clinical PARPi and suggest ways to improve PARPi moving forward.
Figures

References
-
- B. Lüscher, I. Ahel, M. Altmeyer, A. Ashworth, P. Bai, P. Chang, M. Cohen, D. Corda, F. Dantzer, M. D. Daugherty, T. M. Dawson, V. L. Dawson, S. Deindl, A. R. Fehr, K. L. H. Feijs, D. V. Filippov, J. P. Gagné, G. Grimaldi, S. Guettler, N. C. Hoch, M. O. Hottiger, P. Korn, W. L. Kraus, A. Ladurner, L. Lehtiö, A. K. L. Leung, C. J. Lord, A. Mangerich, I. Matic, J. Matthews, G. L. Moldovan, J. Moss, G. Natoli, M. L. Nielsen, M. Niepel, F. Nolte, J. Pascal, B. M. Paschal, K. Pawłowski, G. G. Poirier, S. Smith, G. Timinszky, Z. Q. Wang, J. Yélamos, X. Yu, R. Zaja, M. Ziegler, ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 289, 7399–7410 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

