High Accuracy Prediction of PROTAC Complex Structures
- PMID: 36961978
- PMCID: PMC10240388
- DOI: 10.1021/jacs.2c09387
High Accuracy Prediction of PROTAC Complex Structures
Abstract
The design of PROteolysis-TArgeting Chimeras (PROTACs) requires bringing an E3 ligase into proximity with a target protein to modulate the concentration of the latter through its ubiquitination and degradation. Here, we present a method for generating high-accuracy structural models of E3 ligase-PROTAC-target protein ternary complexes. The method is dependent on two computational innovations: adding a "silent" convolution term to an efficient protein-protein docking program to eliminate protein poses that do not have acceptable linker conformations and clustering models of multiple PROTACs that use the same E3 ligase and target the same protein. Results show that the largest consensus clusters always have high predictive accuracy and that the ensemble of models can be used to predict the dissociation rate and cooperativity of the ternary complex that relate to the degrading activity of the PROTAC. The method is demonstrated by applications to known PROTAC structures and a blind test involving PROTACs against BRAF mutant V600E. The results confirm that PROTACs function by stabilizing a favorable interaction between the E3 ligase and the target protein but do not necessarily exploit the most energetically favorable geometry for interaction between the proteins.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

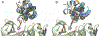
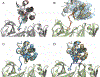
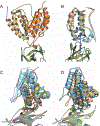


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

