Implementation of genomic surveillance of SARS-CoV-2 in the Caribbean: Lessons learned for sustainability in resource-limited settings
- PMID: 36963002
- PMCID: PMC10022082
- DOI: 10.1371/journal.pgph.0001455
Implementation of genomic surveillance of SARS-CoV-2 in the Caribbean: Lessons learned for sustainability in resource-limited settings
Erratum in
-
Correction: Implementation of Genomic Surveillance of SARS-CoV-2 in the Caribbean: Lessons learned for sustainability in resource-limited settings.PLOS Glob Public Health. 2023 Sep 11;3(9):e0002393. doi: 10.1371/journal.pgph.0002393. eCollection 2023. PLOS Glob Public Health. 2023. PMID: 37695757 Free PMC article.
Abstract
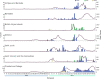
The COVID-19 pandemic highlighted the importance of global genomic surveillance to monitor the emergence and spread of SARS-CoV-2 variants and inform public health decision-making. Until December 2020 there was minimal capacity for viral genomic surveillance in most Caribbean countries. To overcome this constraint, the COVID-19: Infectious disease Molecular epidemiology for PAthogen Control & Tracking (COVID-19 IMPACT) project was implemented to establish rapid SARS-CoV-2 whole genome nanopore sequencing at The University of the West Indies (UWI) in Trinidad and Tobago (T&T) and provide needed SARS-CoV-2 sequencing services for T&T and other Caribbean Public Health Agency Member States (CMS). Using the Oxford Nanopore Technologies MinION sequencing platform and ARTIC network sequencing protocols and bioinformatics pipeline, a total of 3610 SARS-CoV-2 positive RNA samples, received from 17 CMS, were sequenced in-situ during the period December 5th 2020 to December 31st 2021. Ninety-one Pango lineages, including those of five variants of concern (VOC), were identified. Genetic analysis revealed at least 260 introductions to the CMS from other global regions. For each of the 17 CMS, the percentage of reported COVID-19 cases sequenced by the COVID-19 IMPACT laboratory ranged from 0·02% to 3·80% (median = 1·12%). Sequences submitted to GISAID by our study represented 73·3% of all SARS-CoV-2 sequences from the 17 CMS available on the database up to December 31st 2021. Increased staffing, process and infrastructural improvement over the course of the project helped reduce turnaround times for reporting to originating institutions and sequence uploads to GISAID. Insights from our genomic surveillance network in the Caribbean region directly influenced non-pharmaceutical countermeasures in the CMS countries. However, limited availability of associated surveillance and clinical data made it challenging to contextualise the observed SARS-CoV-2 diversity and evolution, highlighting the need for development of infrastructure for collecting and integrating genomic sequencing data and sample-associated metadata.
Copyright: © 2023 Sahadeo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- World Health Organization. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. 2021. Licence: CC BY-NC-SA 3.0 IGO.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
