The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis
- PMID: 36964127
- PMCID: PMC10039029
- DOI: 10.1038/s41467-023-37252-y
The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis
Abstract
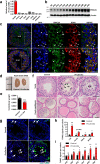
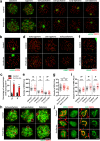
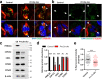
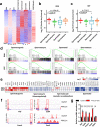
N6-methyladenosine (m6A) and its reader proteins YTHDC1, YTHDC2, and YTHDF2 have been shown to exert essential functions during spermatogenesis. However, much remains unknown about m6A regulation mechanisms and the functions of specific readers during the meiotic cell cycle. Here, we show that the m6A reader Proline rich coiled-coil 2A (PRRC2A) is essential for male fertility. Germ cell-specific knockout of Prrc2a causes XY asynapsis and impaired meiotic sex chromosome inactivation in late-prophase spermatocytes. Moreover, PRRC2A-null spermatocytes exhibit delayed metaphase entry, chromosome misalignment, and spindle disorganization at metaphase I and are finally arrested at this stage. Sequencing data reveal that PRRC2A decreases the RNA abundance or improves the translation efficiency of targeting transcripts. Specifically, PRRC2A recognizes spermatogonia-specific transcripts and downregulates their RNA abundance to maintain the spermatocyte expression pattern during the meiosis prophase. For genes involved in meiotic cell division, PRRC2A improves the translation efficiency of their transcripts. Further, co-immunoprecipitation data show that PRRC2A interacts with several proteins regulating mRNA metabolism or translation (YBX1, YBX2, PABPC1, FXR1, and EIF4G3). Our study reveals post-transcriptional functions of PRRC2A and demonstrates its critical role in the completion of meiosis I in spermatogenesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. doi: 10.1016/S0021-9258(17)32497-3. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

