Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality
- PMID: 36964962
- PMCID: PMC10281603
- DOI: 10.1111/pbi.14042
Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality
Abstract
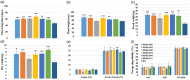
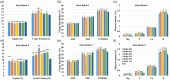
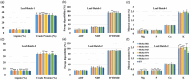
Alfalfa (Medicago sativa L.) is a perennial flowering plant in the legume family that is widely cultivated as a forage crop for its high yield, forage quality and related agricultural and economic benefits. Alfalfa is a photoperiod sensitive long-day (LD) plant that can accomplish its vegetative and reproductive phases in a short period of time. However, rapid flowering can compromise forage biomass yield and quality. Here, we attempted to delay flowering in alfalfa using multiplex CRISPR/Cas9-mediated mutagenesis of FLOWERING LOCUS Ta1 (MsFTa1), a key floral integrator and activator gene. Four guide RNAs (gRNAs) were designed and clustered in a polycistronic tRNA-gRNA system and introduced into alfalfa by Agrobacterium-mediated transformation. Ninety-six putative mutant lines were identified by gene sequencing and characterized for delayed flowering time and related desirable agronomic traits. Phenotype assessment of flowering time under LD conditions identified 22 independent mutant lines with delayed flowering compared to the control. Six independent Msfta1 lines containing mutations in all four copies of MsFTa1 accumulated significantly higher forage biomass yield, with increases of up to 78% in fresh weight and 76% in dry weight compared to controls. Depending on the harvesting schemes, many of these lines also had reduced lignin, acid detergent fibre (ADF) and neutral detergent fibre (NDF) content and significantly higher crude protein (CP) and mineral contents compared to control plants, especially in the stems. These CRISPR/Cas9-edited Msfta1 mutants could be introduced in alfalfa breeding programmes to generate elite transgene-free alfalfa cultivars with improved forage biomass yield and quality.
Keywords: FLOWERING LOCUS T; CRISPR/Cas9; alfalfa; biomass; genome editing; quality.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Aung, B. , Gruber, M.Y. , Amyot, L. , Omari, K. , Bertrand, A. and Hannoufa, A. (2015) MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. - PubMed
-
- Ball, D.M. , Collins, M. , Lacefield, G.D. , Martin, N.P. , Mertens, D.A. , Olson, K.E. et al. (2001) Understanding Forage Quality. Park Ridge, IL: American Farm Bureau Federation Publication 1‐01.
-
- Bao, Q. , Wolabu, T.W. , Zhang, Q. , Zhang, T. , Liu, Z. , Sun, J. and Wang, Z.Y. (2022) Application of CRISPR/Cas9 technology in forages. Grassland Res. 1, 244–251.
-
- Bhattarai, K. , Rajasekar, S. , Dixon, R.A. and Monteros, M.J. (2018) Agronomic performance and lignin content of HCT down‐regulated alfalfa (Medicago sativa L.). Bioenergy Res. 11, 505–515.
-
- Bucher, S.F. and Römermann, C. (2021) The timing of leaf senescence relates to flowering phenology and functional traits in 17 herbaceous species along elevational gradients. J. Ecol. 109, 1537–1548.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

