Spatiotemporal Characterization of Human Early Intervertebral Disc Formation at Single-Cell Resolution
- PMID: 36965031
- PMCID: PMC10190614
- DOI: 10.1002/advs.202206296
Spatiotemporal Characterization of Human Early Intervertebral Disc Formation at Single-Cell Resolution
Abstract
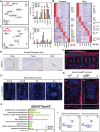
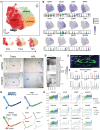
The intervertebral disc (IVD) acts as a fibrocartilaginous joint to anchor adjacent vertebrae. Although several studies have demonstrated the cellular heterogeneity of adult mature IVDs, a single-cell transcriptomic atlas mapping early IVD formation is still lacking. Here, the authors generate a spatiotemporal and single cell-based transcriptomic atlas of human IVD formation at the embryonic stage and a comparative mouse transcript landscape. They identify two novel human notochord (NC)/nucleus pulposus (NP) clusters, SRY-box transcription factor 10 (SOX10)+ and cathepsin K (CTSK)+ , that are distributed in the early and late stages of IVD formation and they are validated by lineage tracing experiments in mice. Matrisome NC/NP clusters, T-box transcription factor T (TBXT)+ and CTSK+ , are responsible for the extracellular matrix homeostasis. The IVD atlas suggests that a subcluster of the vertebral chondrocyte subcluster might give rise to an inner annulus fibrosus of chondrogenic origin, while the fibroblastic outer annulus fibrosus preferentially expresseds transgelin and fibromodulin . Through analyzing intercellular crosstalk, the authors further find that notochordal secreted phosphoprotein 1 (SPP1) is a novel cue in the IVD microenvironment, and it is associated with IVD development and degeneration. In conclusion, the single-cell transcriptomic atlas will be leveraged to develop preventative and regenerative strategies for IVD degeneration.
Keywords: annulus fibrosus; intervertebral disc formation; notochord; nucleus pulposus; single-cell RNA sequencing.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Boos N., Weissbach S., Rohrbach H., Weiler C., Spratt K. F., Nerlich A. G., Spine 2002, 27, 2631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
